Figure 2: Sensitivity and accuracy of cLC-SRM for multiplexed quantification of EGFR/MAPK pathway proteins.
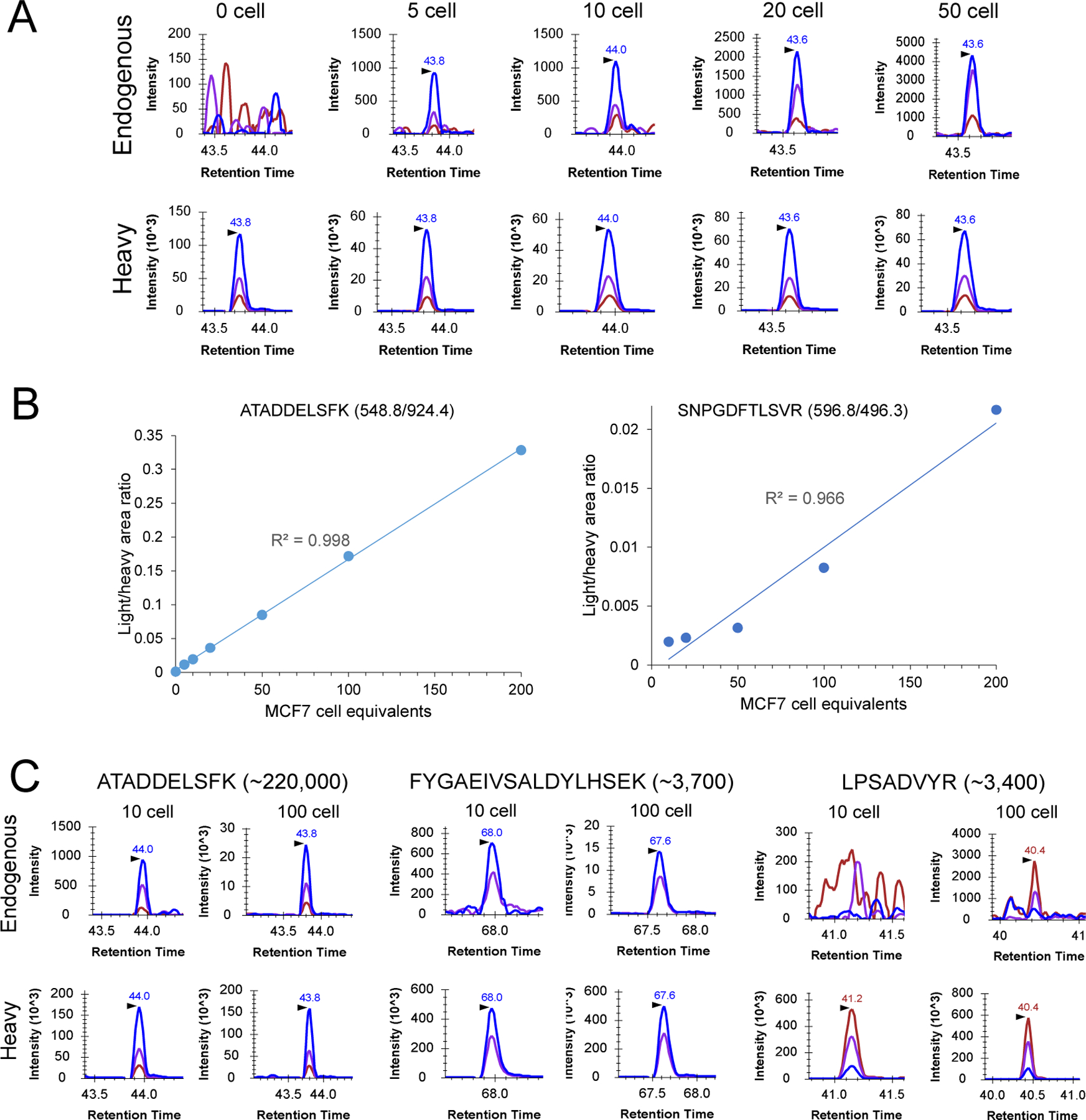
(A) Extracted ion chromatograms (XICs) of transitions monitored for ATADDELSFK derived from GRB2 at different numbers of MCF7 cell equivalents: 548.8/924.4 (blue), 548.8/853.4 (purple), 548.8/738.4 (chestnut). (B) Calibration curves for quantifying high abundance GRB2 and moderate-abundance PTPN11 with the use of the best responsive interference-free transitions, ATADDELSFK (548.8/924.4) for GRB2 and SNPGDFTLSVR (596.9/496.3) for PTPN11. Three and two SRM replicates were performed for 0–10 and 20–200 MCF7 cell equivalents, respectively. (C) Comparison of SRM signal between 10 and 100 MCF7 cells sorted by FACS. Each sample consists of two biological replicates with the addition of ~30 fmol of heavy peptide standards per replicate. XICs of transitions monitored for ATADDELSFK derived from GRB2: 548.8/924.4 (blue), 548.8/853.4 (purple), 548.8/738.4 (chestnut); XICs of transitions monitored for FYGAEIVSALDYLHSEK derived from AKT1: 648.0/897.9 (blue), 648.0/816.4 (purple), 648.0/283.1 (chestnut); XICs of transitions monitored for LPSADVYR derived from SOS1: 460.7/807.4 (blue), 460.7/710.3 (purple), 460.7/404.2 (chestnut).
