Figure 6. Predicting new antibiotic candidates from unprecedented chemical libraries.
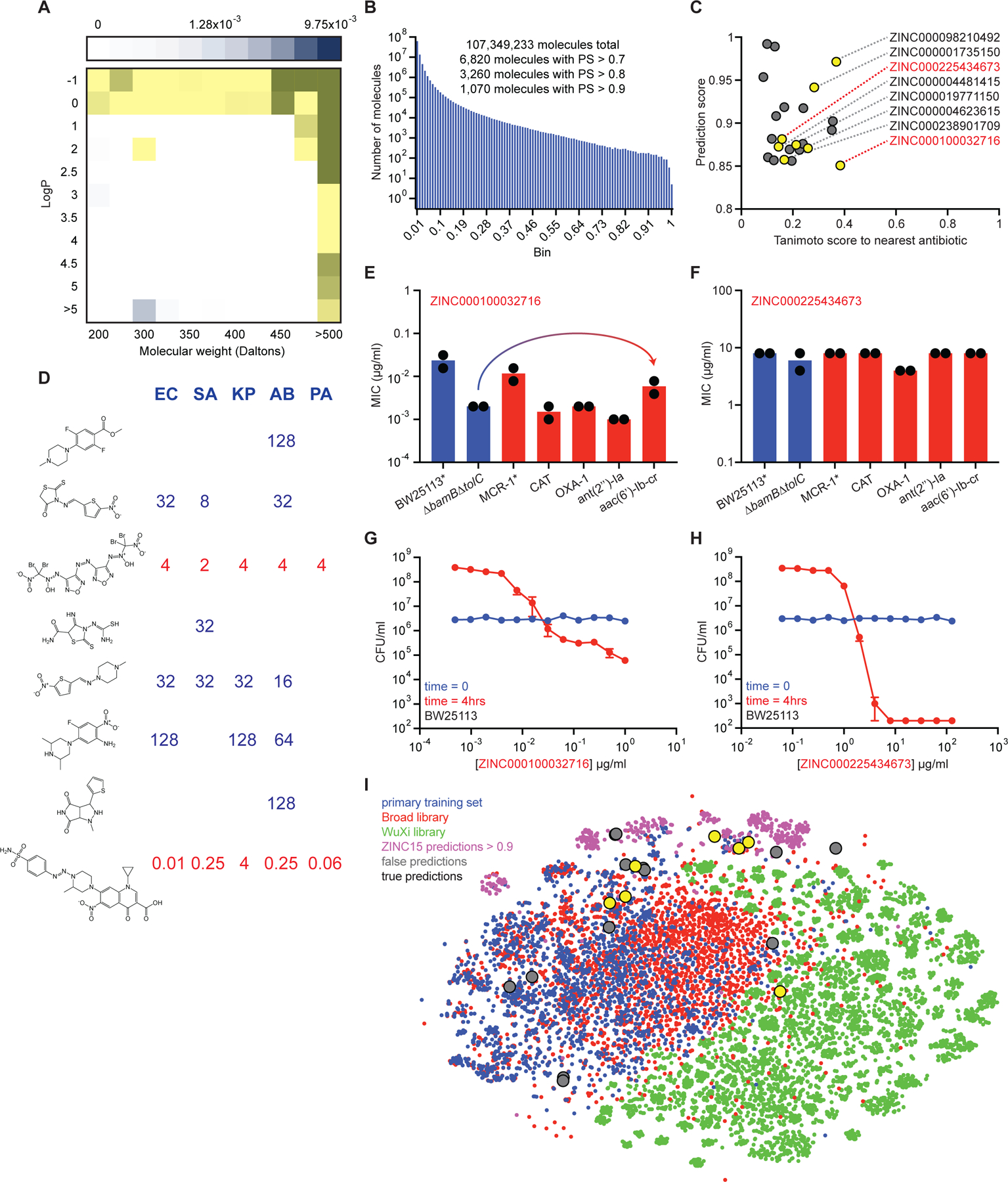
(A) Tranches of the ZINC15 database colored based on the proportion of hits from the original training dataset of 2,335 molecules within each tranche. Darker blue tranches have a higher proportion of molecules that are growth inhibitory against E. coli. Yellow tranches are those selected for predictions. (B) Histogram showing the number of ZINC15 molecules from selected tranches within a corresponding prediction score range. (C) Prediction scores and Tanimoto nearest neighbour antibiotic scores of the 23 predictions that were empirically tested for growth inhibition. Yellow circles represent those molecules that displayed detectable growth inhibition of at least one pathogen. Grey circles represent inactive molecules. ZINC numbers of active molecules are shown on the right. (D) MIC values (µg/ml) of the eight active predictions from the ZINC15 database against E. coli (EC), S. aureus (SA), K. pneumoniae (KP), A. baumannii (AB), and P. aeruginosa (PA). Blank regions represent no detectable growth inhibition at 128 µg/ml. Structures are shown in the same order (top to bottom) as their corresponding ZINC numbers in (C). (E) MIC of ZINC000100032716 against E. coli strains harboring a range of antibiotic-resistance determinants. The mcr-1 gene was expressed in E. coli BW25113. All other resistance genes were expressed in E. coli BW25113 ∆bamB∆tolC. Experiments were conducted with two biological replicates. Note the minor increase in MIC in the presence of aac(6’)-Ib-cr. (F) Same as (E) except using ZINC000225434673. (G) Killing of E. coli in LB media in the presence of varying concentrations of ZINC000100032716 after 0 hr (blue) and 4 hr (red). The initial cell density is ~106 CFU/ml. Shown is the mean of two biological replicates. Bars denote absolute error. (H) Same as (G) except using ZINC000225434673. (I) t-SNE of all molecules from the primary training dataset (blue), the Drug Repurposing Hub (red), the WuXi anti-tuberculosis library (green), the ZINC15 molecules with prediction scores >0.9 (pink), false positive predictions (grey), and true positive predictions (yellow). See also Figure S5, Table S5–S7.
