Figure 2: Generation and characterization of the mouse Adamts7 E373Q catalytic mutant allele.
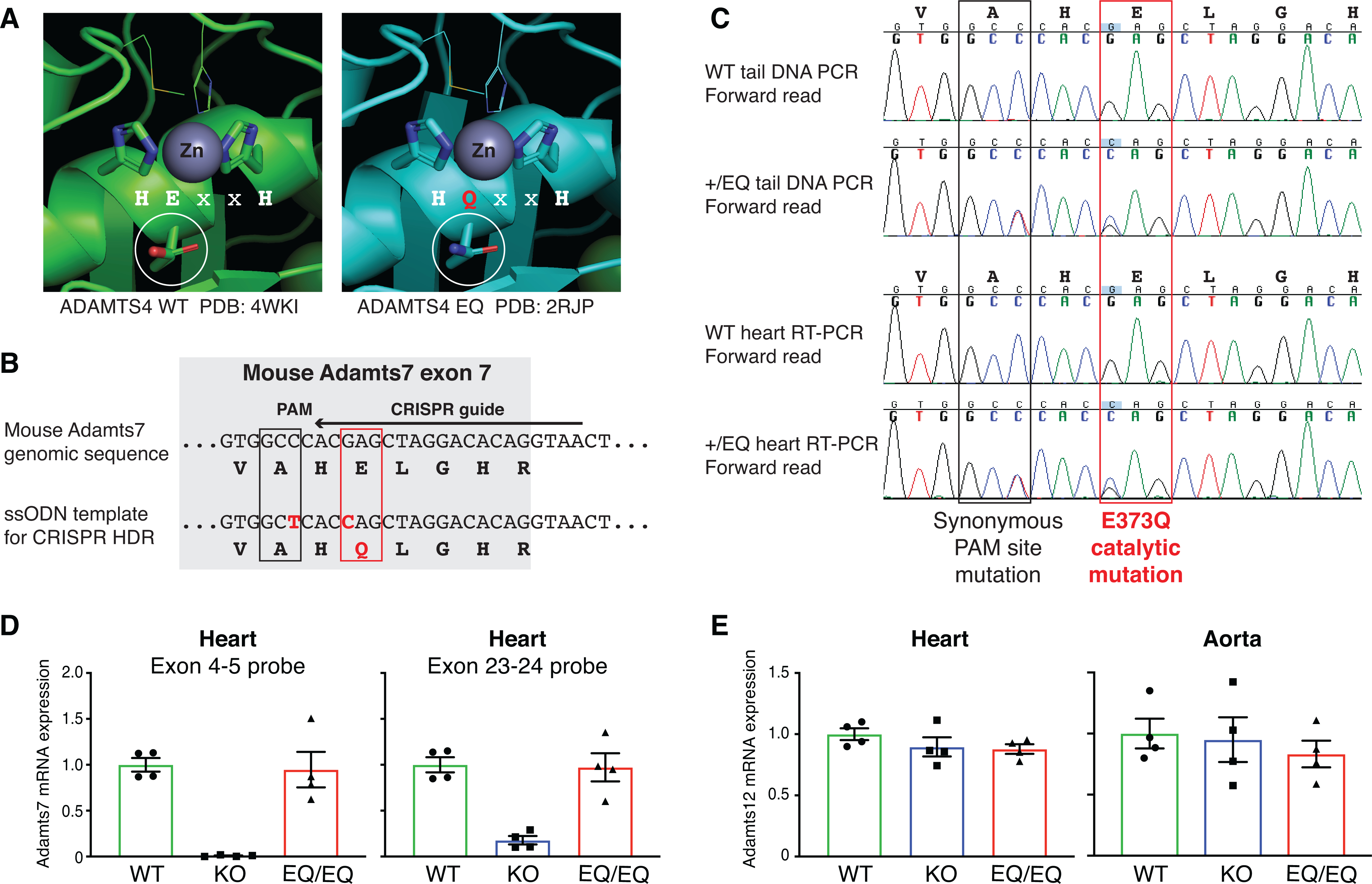
A, Structural context of the ADAMTS7 HExxH to HQxxH catalytic mutation. Crystal structures from ADAMTS4 WT (PDB:4WKI)33 and ADAMTS4 EQ (PDB: 2RJP)34 were aligned and annotated in PyMOL to highlight the catalytic residues and visualize the Zinc metal in the active site. The E to Q substitution preserves the tertiary structure of ADAMTS4 and the residues “VAHELGH” in the active site are conserved in ADAMTS7. B, Schematic of the Adamts7 E373Q catalytic mutant allele within exon 7. To generate the mutant allele, c. 1117G->C (p. E373Q) mutation at the Adamts7 catalytic domain and c. 1113C->T (p. A371A) to disrupt the PAM site were induced by CRISPR Homology-Directed Repair (HDR). C, Sanger sequencing of genomic tail DNA PCR and heart mRNA RT-PCR from WT and heterozygous +/E373Q mice. Representative forward reads show no evidence of allelic expression imbalance at the two nucleotide substitutions. D, Adamts7 mRNA expression level in the heart from WT, Adamts7 −/− (KO) and homozygous E373Q/E373Q (EQ/EQ) mice were measured by real time quantitative polymerase chain reaction (qPCR) using 2 TaqMan probe sets (exon 4–5 boundary and exon 23–24 boundary). n=4 per group. E, Adamts12 mRNA expression levels were analyzed by real time qPCR in the heart and aorta harvested from WT, KO and EQ/EQ mice. n=4 per group. A Kruskal-Wallis test with Dunn’s multiple comparison test was applied to D and E.
