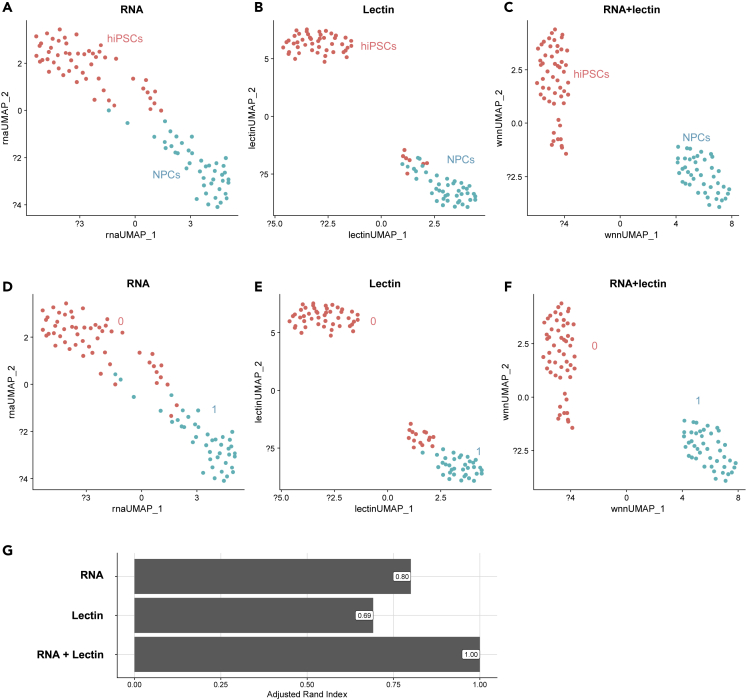
Figure 7.
Dimensional reduction and clustering using the Seurat workflow
(A–C) UMAP visualization based on (A) only the scRNA-seq data and (B) only the scGlycan-seq, and (C) both scRNA-seq and scGlycan-seq (scGR-seq) data of hiPSCs (n = 53, red) and NPCs (n = 43, green). UMAP computation was performed based on the weighted nearest neighbor graph that integrates the RNA and lectin modalities.
(D–F) Same UMAP visualization as (A-C) but single cells are colored by the clustering results based on (D) only scRNA-seq data, (E) only scGlycan-seq data, and (F) both scRNA-seq and scGlyca-seq (scGR-seq) data using a shared nearest neighbor (SNN) modularity optimization-based clustering algorithm.
(G) Bar plots showing adjusted Rand index as a measure of the similarity of the clustering in (D), (E), and (F) to the cell type annotation (A–C).
See also Figures S9–S13.