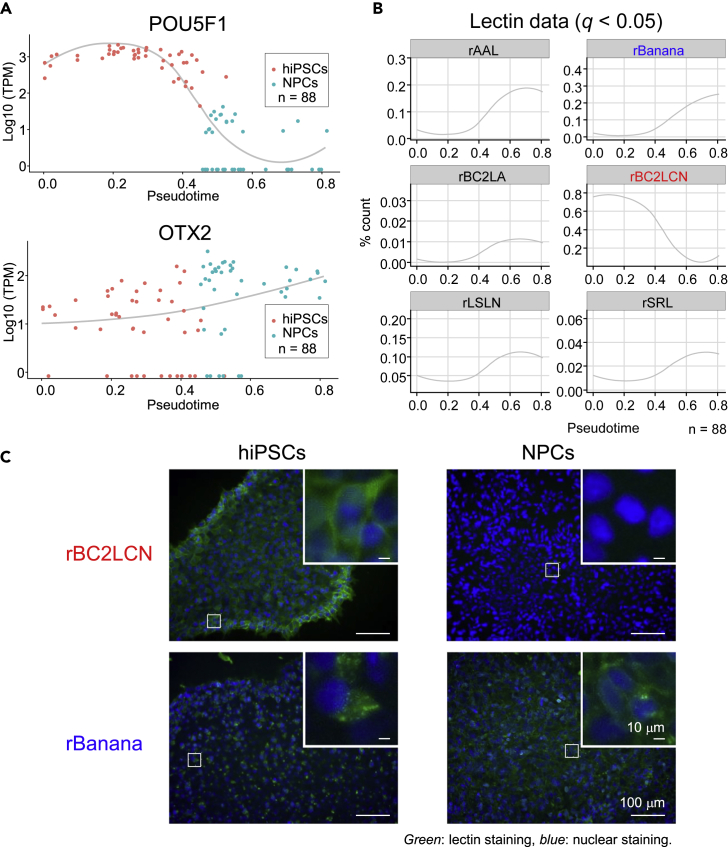
Figure 9.
Psuedotime analysis of mRNA and lectin signals
(A) Expression profile of the hiPSC marker (POU5F1) and NPC marker (OTX2) genes. The x axis represents the pseudotime inferred by slingshot using mRNA data. The red and blue points indicate hiPSCs and NPCs, respectively. The curve represents smoothed conditional means calculated by the LOESS (locally estimated scatterplot smoothing) method.
(B) Dynamically regulated lectins along the pseudotime. The pseudotime is the same as in (A). A generalized additive model was fitted to the lectin signals to search for dynamically regulated lectins (q < 0.05 after the multiple testing correction using the Benjamini and Hochberg method). The red and blue points indicate hiPSCs and NPCs, respectively. The curve represents smoothed conditional means calculated by the LOESS method.
(C) Fluorescence staining of hiPSCs and hiPSC-derived NPCs by rBC2LCN and rBanana. hiPSCs and hiPSC-derived NPCs (17-day differentiation) were fixed with 4% paraformaldehyde at room temperature for 20 min. After blocking with PBS containing 1% BSA at room temperature for 30 min, cells were incubated with 1 μg/mL of fluorescently labeled rBC2LCN and rBanana for 1 hr at room temperature and observed by fluorescence microscopy. The nucleus was stained with Hoechst 33,342 (x 1,000) at room temperature for 5 min. Insets show high magnification of selected fields. Nucleus: blue. lectin: green. Scale bar: 100 μm or 10 μm (insets).
See also Figures S16 and S17.