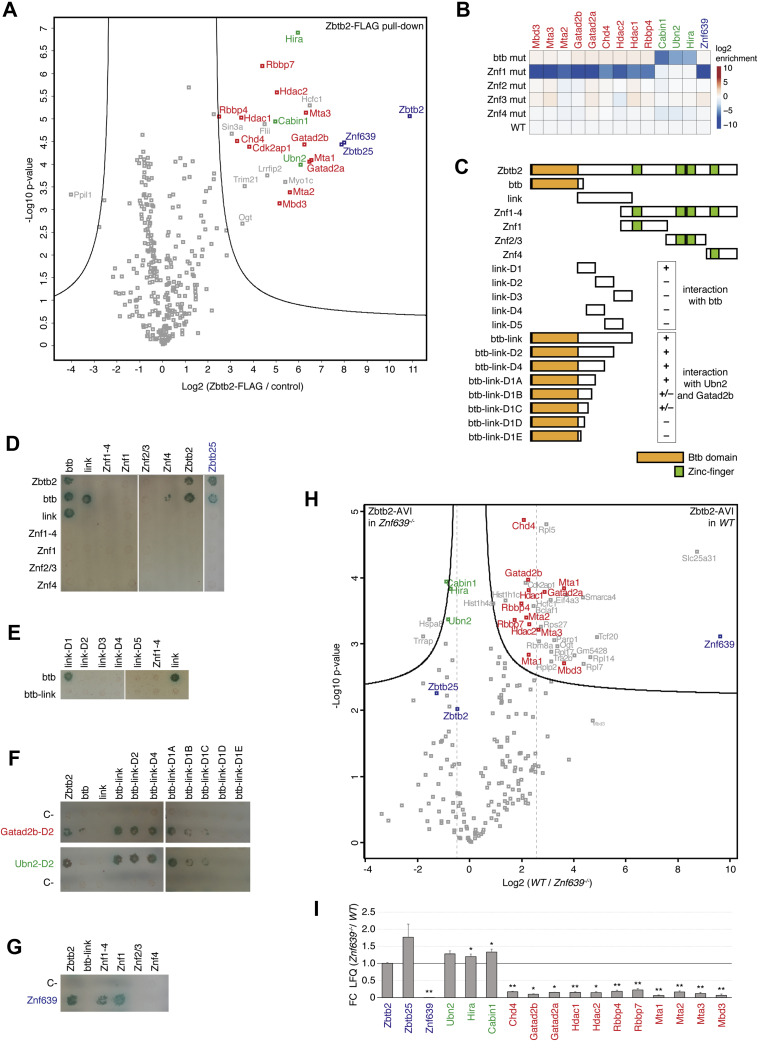
Figure 2.
An extended BTB domain binds UBN2 and GATAD2B; NuRD interaction is stabilized by ZNF639.A, volcano plot of protein enrichments in anti-FLAG AP-MS from mESCs expressing endogenous ZBTB2-3xFLAG compared with parental untargeted mESCs. ZBTB2 and partner TFs are indicated in blue, NuRD subunits in red, and HiRA subunits in green. B, bait-normalized log2 fold-enrichment of ZNF639, HiRA subunits, and NuRD subunits in AP-MS from mESCs overexpressing ZBTB2-AVI alleles with indicated mutations compared with overexpressing WT ZBTB2-AVI. All baits (Table S4) were induced with DOX in Zbtb2−/−Zbtb25−/− mESCs. C, diagram of ZBTB2 constructs used for Y2H analysis; plus and minus indicate positive and negative interactions as in panels E and F. D–G, colony growth on SD-Ade/-His/-Trp/-Leu/+X-alpha-Gal/+Aureobasidin A (QDOXA) plates of strains expressing indicated proteins. Bait constructs are vertical (D and E) and horizontal (F and G), and prey constructs are horizontal (D and E) and vertical (F and G). H, volcano plot of protein enrichments in AP-MS of overexpressed ZBTB2-AVI in WT compared with Znf639−/− mESCs. Color code as in Figure 2A. Dashed lines highlight the difference in NuRD subunit abundance relative to the bait ZBTB2. I, bait-normalized interaction changes as in panel H. ∗p-value < 0.01 and ∗∗p-value < 0.001. AP-MS, affinity purification–MS; BTB, broad-complex, tramtrack, and bric-a-brac; DOX, doxycycline; HiRA, histone regulator A; mESCs, mouse embryonic stem cells; NuRD, nucleosome remodeling and deacetylase; TFs, transcription factors; Y2H, yeast-2-hybrid.