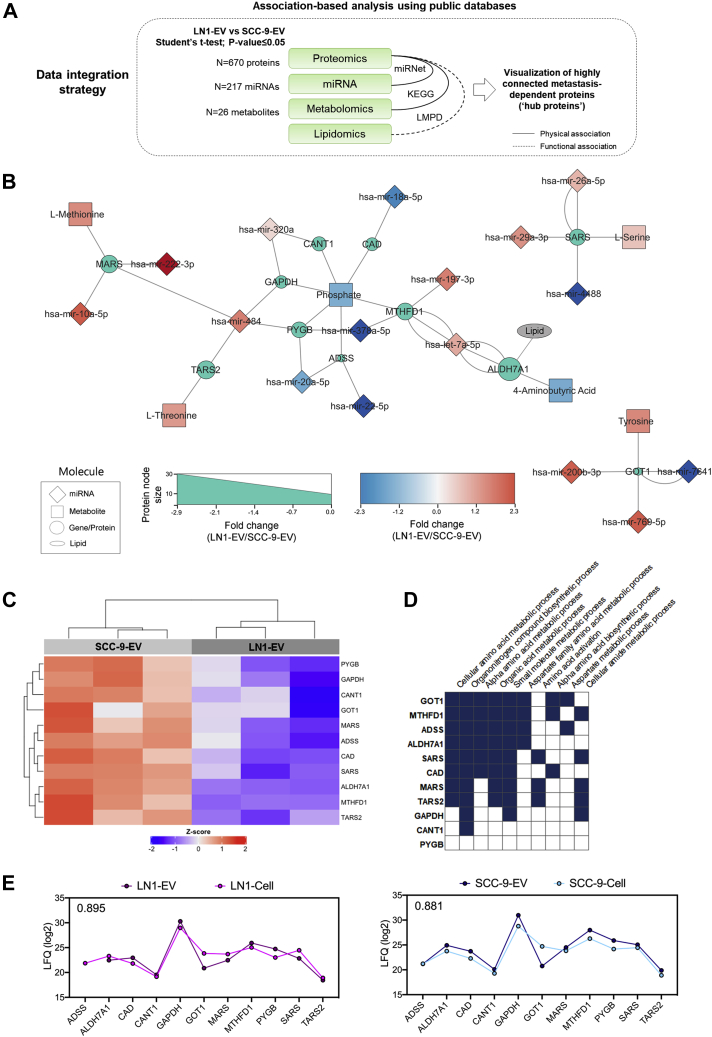
Fig. 4.
Multi-omics integrative analysis of proteomics, miRNA, metabolomics, and lipidomics content from SCC-9 and LN1-derived EVs. The integration of differentially abundant molecules from EVs (Student's t test; p-value ≤ 0.05; LN1 EVs versus SCC-9 EVs) based on associations (A) showed as a set of 11 ‘hub proteins’ that interact with specific miRNAs or metabolites, as well as the presence of the lipid-associated protein ALDH7A1 (B). All ‘hub proteins’ are downregulated in LN1 EVs when compared with SCC-9 EVs (C) and may modulate specific biological processes associated with metabolism (D). The heat map was generated using the Euclidean distance and complete linkage method (n = 11 proteins). The correlation between protein abundance in cells and EVs was calculated for the 11 selected proteins considering SCC-9 and LN1 protein intensities (E). The Pearson coefficient is shown in the upper left corner of the graphs. EVs, extracellular vesicles.