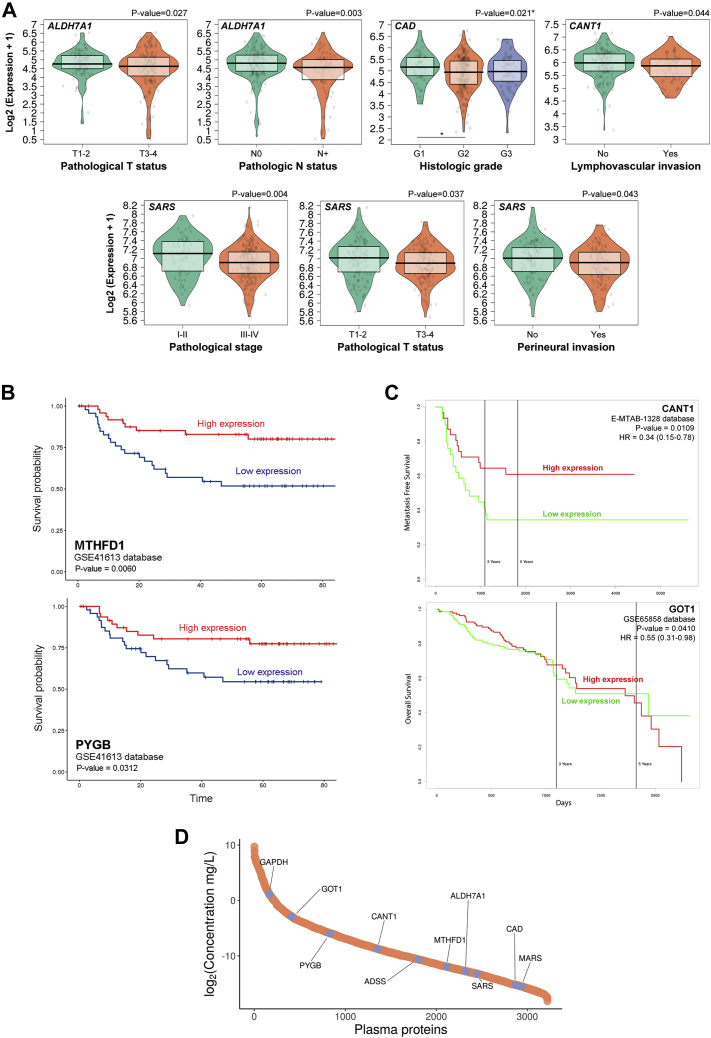
Fig. 5.
Characterization of prognostic markers in HNSCC using public databases. TCGA, GSE41613, GSE65858, and MTAB-1328 repositories were used to determine proteins from the multi-omics integrative analysis associated with clinical and pathological features in patients with HNSCC. Using TCGA database, the gene expression pattern of a group of hub proteins was significantly associated with clinical features (A; ALDH7A1, CAD, CANT1, SARS, p-value ≤ 0.05). MTHFD1 and PYGB downregulation was linked to poor overall survival in GSE41613 database using HNCDB tool (B), whereas CANT1 and GOT1 low transcript levels were associated with poor overall survival and metastasis-free survival in GSE-65858 and E-MTAB-1328 databases, respectively (C) (p-value ≤ 0.05). The concentration of plasma proteins from healthy humans was retrieved for the 11 ‘hub proteins’ from MS experiments available in The Human Plasma Proteome and represented in a dynamic range plot (D). The proteins of interest are highlighted in the graph. HNCDB, Head and Neck Cancer Database; HNSCC, head and neck squamous cell carcinoma; N0, patient negative for lymph node metastasis considering pathological staging; N+, patient positive for lymph node metastasis considering pathological staging; TCGA, The Cancer Genome Atlas.