Abstract
Venomous snakebite is a neglected tropical disease that annually leads to hundreds of thousands of deaths or long-term physical and mental ailments across the developing world. Insufficient data on spatial variation in snakebite risk, incidence, human vulnerability, and accessibility of medical treatment contribute substantially to ineffective on-ground management. There is an urgent need to collect data, fill knowledge gaps and address on-ground management problems. The use of novel, and transdisciplinary approaches that take advantage of recent advances in spatio-temporal models, ‘big data’, high performance computing, and fine-scale spatial information can add value to snakebite management by strategically improving our understanding and mitigation capacity of snakebite. We review the background and recent advances on the topic of snakebite related geospatial analyses and suggest avenues for priority research that will have practical on-ground applications for snakebite management and mitigation. These include streamlined, targeted data collection on snake distributions, snakebites, envenomings, venom composition, health infrastructure, and antivenom accessibility along with fine-scale models of spatio-temporal variation in snakebite risk and incidence, intraspecific venom variation, and environmental change modifying human exposure. These measures could improve and ‘future-proof’ antivenom production methods, antivenom distribution and stockpiling systems, and human-wildlife conflict management practices, while simultaneously feeding into research on venom evolution, snake taxonomy, ecology, biogeography, and conservation.
Keywords: Snakebite incidence, Envenomings, Neglected tropical diseases, Spatio-temporal epidemiology, Medically relevant snakes, Species distribution models
Highlights
-
•
Many knowledge gaps remain on snake distributions and spatial snakebite variation.
-
•
Targeted data collection and high resolution spatial models are needed.
-
•
Maps of snake distributions, bite incidence, and vulnerable populations are needed.
-
•
Area-specific antivenom delivery requires studies on spatial venom variation.
-
•
Human welfare and snake conservation require spatial management of conflict.
1. Background
Venomous snakebite is recognized as a ‘category A’ neglected tropical disease (NTD) by the World Health Organization (Longbottom et al., 2018; WHO, 2017; Williams et al., 2010; Williams et al., 2011) and disproportionately affects agricultural workers, especially young males in poor rural communities in the developing world (Hansdak et al., 1998 [Nepal]; Harrison et al., 2009 & Mohapatra et al., 2011 [India]; Yates et al., 2010 [Tanzania]; Dehghani et al., 2014 [Iran]; Mendonça-da-Silva et al., 2017 [Brazil]; Ediriweera et al., 2019 [Sri Lanka]). The most heavily affected regions are tropical sub-Saharan Africa, the Indian subcontinent, South-East Asia, and tropical Latin America (Ediriweera et al., 2019; Kasturiratne et al., 2008). Estimates of the number of people affected globally vary greatly: between 1.2 and 5.5 million people are bitten every year, 420,000–2.7 million are envenomed, up to 137,880 die, and a further ~400,000 suffer from resulting long-term medical conditions (Chippaux, 1998; Gutiérrez et al., 2017; Kasturiratne et al., 2008; Mion and Olive, 1997). Despite high snakebite prevalence, substantial knowledge gaps on many components of the issue remain, existing knowledge is often outdated, and, as shown by large ranges in bite [1.2–5.5 million] and envenoming [420,000–2.7 million] estimates provided above, contemporary burden estimates lack precision. Knowledge gaps directly stem from:
-
(i)
historical lack of investment into research on medical conditions that primarily affect the developing world,
-
(ii)
difficulties involved in data collection across remote regions with limited physical accessibility, unstable political conditions, and lack of reliable reporting systems, and
-
(iii)
limited access to and affordability of medical treatment, resulting in poor medical records on the distribution and frequency of snakebite.
The resulting knowledge gaps have clear spatial components, i.e. to effectively distribute antivenoms and manage snakebite more generally, we need to understand the geographic variation of causative processes and their consequences, and identify efficient interventions from a geographical perspective, in addition to addressing the cultural, and financial problems. The main knowledge gaps fit into several broad categories:
1.1. Sparse & heterogeneous data
Firstly, sparse and heterogeneous data on distributions and geographic variation in abundance of medically relevant snake species (Genevieve et al., 2018; Gutiérrez et al., 2013; Yañez-Arenas et al., 2016), exposure of vulnerable human populations to venomous snakes, snakebite frequency (Gutiérrez et al., 2010; Longbottom et al., 2018), and community-based epidemiology (Ediriweera et al., 2016) lead to a lack of knowledge on high risk snakebite areas, and on adequate prioritization for the improvement of access to antivenom and medical facilities or preventive intervention campaigns. This lack of data stands in stark contrast with the potential benefits of using ‘big data’ spatio-temporal modelling approaches to analyze relevant patterns. Whilst rich distribution datasets exist for some snakes, e.g. in the Americas and Europe (Nogueira et al., 2019; Sillero et al., 2014), such data is not complete across all relevant snake species and spatial domains. Additionally, snakebite incidence data are collected by a variety of methods, ranging from community-based randomized surveys to clinical presentations, which makes direct comparisons across geographical areas challenging. Lastly, many aspects of snake biology that could help with predicting the epidemiology of snakebite (abundance, population dynamics, etc.) are understudied (Murray et al., 2020).
1.2. Changing processes
Secondly, our world is changing rapidly due to climate change (IPCC, 2019; O'Connor et al., 2020; Ortiz et al., 2021; Peace, 2020) and human land use change (Hurtt et al., 2020; Li et al., 2017; Ortiz et al., 2021). Both processes affect the spatial use of land by humans and snakes, and consequently their interactions (Ediriweera et al., 2018; Goldstein et al., 2021; Martin et al., 2021; this issue). Predicting how snakebite prevalence and distribution will change is challenging and requires urgent attention to ensure successful snakebite management.
1.3. Antivenom research
Thirdly, the efficacy of available antivenoms and geographic variation thereof is poorly characterized. Because of limited quality control and case studies, it is often unclear which species or populations were used to create each antivenom, how much of the antivenom is required to effectively treat envenomation by each species, and sometimes even if the antivenom effectively neutralizes venom of a certain species at all (Chippaux et al., 1991; Fry et al., 2003; Gutiérrez et al., 2010, 2011; Saravia et al., 2002; Warrell, 1997; Williams et al., 2011). These issues are exacerbated by substantial intraspecific venom variation (Casewell et al., 2014, 2020; Currier et al., 2010; Daltry et al., 1996; Pla et al., 2019; Senji Laxme et al., 2021a, 2021b), and limited knowledge on the geographic distribution of different intraspecific ‘venom lineages’.
1.4. On ground measures
Lastly, there is limited financial investment in antivenom improvements, availability of protective equipment, and access to high quality medical treatment. Victims are often hours away from medical facilities and cannot afford treatment, and often seek local healers instead of western medicine (Ediriweera et al., 2017; Newman et al., 1997). Additionally, farmers often tend to fields barefoot (particularly rice), and dwellings generally offer limited protection from wildlife (Harrison et al., 2009). These factors highlight the urgent need for stockpiles of free, high-quality antivenoms in strategic locations along with provision of protective equipment (WHO, 2019). Encouraging such measures requires accompanying community engagement and education campaigns (WHO, 2019), to build community knowledge and appreciation for the importance of snakebite prevention, adequate first aid, and attendance of approved medical facilities.
In response to the impact of snakebite on health and economies in the developing world (Habib and Brown, 2018; Harrison and Gutiérrez, 2016; Kasturiratne et al., 2008, 2017), WHO has compiled new guidelines for antivenom production and testing (WHO, 2010a, 2018) and plans to stockpile antivenoms at key facilities to alleviate and manage the issue (WHO, 2019). Such efforts would benefit from filling the above knowledge gaps.
Over the last decade, spatial analytical methods and availability of high resolution, high quality spatial datasets have increased immensely, along with advancements in ‘big data’ processing capacities, high resolution satellite imagery, and access to high performance computing facilities. Many tasks that would have been computationally prohibitive 10 years ago have become feasible in recent years. While many traditional spatial analytics prove useful for the analysis of spatial patterns in snakebite epidemiology, more advanced approaches to solving the World's problems require revaluation at a frequent rate; snakebite management is a good example of this. Numerous new approaches to some of the spatial challenges outlined above, or similar ones in different fields, have been developed and successfully applied to varying regions of the world. This review aims to provide a transdisciplinary summary of recent advances in managing the global snakebite crisis from a spatial perspective using novel spatio-temporal modelling and ‘big data’ approaches.
Because the relevant literature and knowledge gaps span a broad range of topics and sub-topics, we review them in individual sections. First we discuss the sparsity of data on snake distributions [section 2], and how the typically conservation related field of human-wildlife conflict can lead to a unique, transdisciplinary scenario akin to, but distinct from traditional epidemiology [section 3]. We then elaborate on how an improved understanding of snake biology [section 4] and spatio-temporal patterns in snakebite incidence [section 5] is needed to address the global snakebite crises. This is followed by a review of how human populations become particularly vulnerable to the medical consequences of snakebite and how such vulnerability can be mitigated by spatial optimization of medical resource allocation [section 6]. The penultimate section [section 7] synthesizes the dynamic nature of snakebite epidemiology by describing how climate change and land use change need to be incorporated into analyses to keep mitigation efforts up-to-date. Lastly [section 8], we discuss the geographic aspects of antivenom distribution and development, which is quite distinct from the previous sections and ties snake biology and on-ground snakebite management into medical pharmacology. Throughout, we provide table overviews of key literature, give details of where ‘big data’ approaches are currently hindered by insufficient existing data, and suggest how remaining knowledge gaps could be closed to resolve practical challenges in snakebite management.
2. Back to basics: improving our knowledge of snake distributions
Despite the enormous burden snakebite causes every year, our understanding of some basic features of the issue remains limited. WHO maintains a list of medically relevant snakes (WHO, 2018); updated annually, David Williams pers. com.), their known distributions (WHO, 2010b); previously updated infrequently but soon biannually), and their categorization into class 1 (highest medical importance) or 2 (secondary medical importance; see https://apps.who.int/bloodproducts/snakeantivenoms/database/), depending on the impact they cause in any given country (WHO 2018). Taxonomic revisions of snake taxa warrant a rigorous and continuous review process, which is currently under development in form of an interactive online WHO database (David Williams, pers. Com). Establishing accurate distribution maps of snakes is often hampered by surprising data sparsity. Even category 1 species that contribute immensely to the global burden of snakebite sometimes have few verified geographic occurrence localities, and data availability for range restricted, threatened or rarer taxa is much worse (Fig. 1). This showcases the dual need of distribution information for epidemiology as well as for conservation management. It is noteworthy that WHO listed species only include those that contribute substantially to the annual snakebite burden - snakes which cause occasional bites or less severe symptoms are often even more data deficient. Snake distribution estimates are usually based on limited scientific literature and expert opinion. Range estimates are provided by different databases (such as the latest WHO distribution estimate (Longbottom et al., 2018; WHO, 2010b), ‘the Global Assessment of Reptile Distributions’ (Meiri et al., 2017; Roll et al., 2017) and ‘RepFocus’ (Midtgaard, 2021), which often disagree (Fig. 1). Such discrepancies stem from differences in occurrence records used and from different interpretations of what best defines the habitat of a species (boundaries may be drawn subjectively based on similarities in vegetation or altitude), factors which need to be resolved urgently.
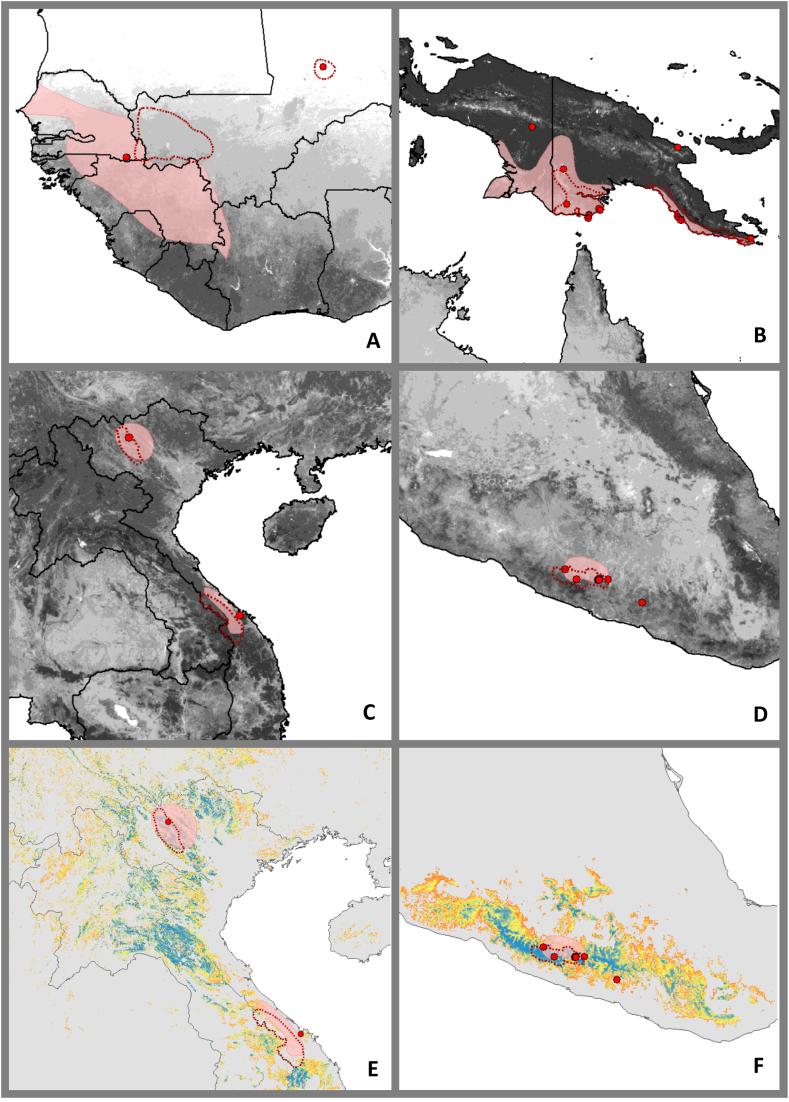
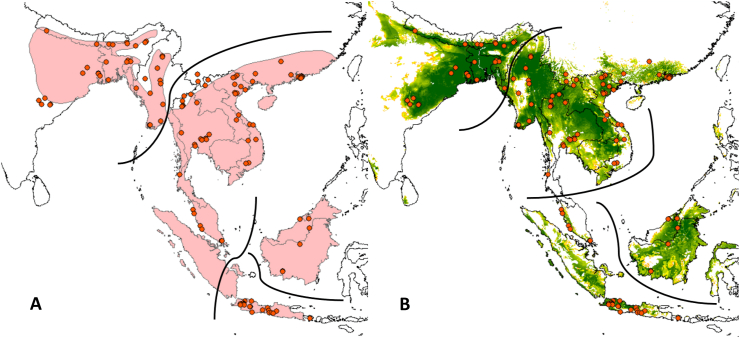
Fig. 1.
WHO (pink shaded area) and GARD (red dotted outlines) distribution estimates, and known occurrences (red dots) for medically relevant snake species of conservation concern (IUCN 2020) from category 1 Echis jogeri [A; data deficient] and Bungarus slowinskii [C & E; vulnerable] and category 2 Pseudechis papuanus [B; data deficient] and Mixcoatlus barbouri [D & F; endangered], showcasing how snakes often have limited distribution data and varying distribution estimates. ENMs for B. slowinski (E) and M. barbouri F) improve distribution estimates (blue = more suitable; data for models was combined with closely related, ecologically similar sister species B. bungaroides and M. browni, respectively, to achieve minimum data requirements for models). Note that suitable habitat may be unreachable by a species or may be occupied by closely related or competing taxa. Background in A-D shows mean vegetation greenness (fraction photosynthetic active radiation; https://land.copernicus.eu/global/products/fapar) with greener shown as darker shades of grey.
Point (fine-scale) occurrence data lies at the core of most distribution estimates. These data come from a combination of different sources including primary literature records, museum records, and other observations, and are often collated in public and private databases. Some frequently used public databases are global platforms such as the Global Biodiversity Information Facility (GBIF, 2021), USGSs Biodiversity Information Serving Our Nation (BISON, 2021), Biocollections (iDigBio, 2021), VertNet (2021), Arctos (2021), as well as country specific platforms (e.g., the Atlas of Living Australia (ALA, 2021), and a growing number of citizen science platforms such as iNaturalist (iNaturalist, 2021) or HerpMapper (2021). The ever-growing number of publicly accessible databases presents new opportunities for biodiversity research, although biodiversity data is unfortunately typically spatially and temporally biased (Boakes et al., 2010) towards developed regions, i.e., the USA, Europe, and Australia (Peterson, 2014), and towards accessible areas within regions (Ficetola et al., 2013; Piccolo et al., 2020).
Data from taxonomically reliable sources such as museum records and scientific literature has its obvious advantages: often they relate to voucher specimen or DNA samples, which enable re-examination to verify identification or re-attribution after taxonomic revisions. However, enormous advances in data processing capacities over the last decade, combined with the ever-growing number of mobile phone devices with cameras used by the general public even in the developing world, present a promising opportunity to fill data gaps without the need for time consuming and costly fieldwork by experts. For example, iNaturalist has a mobile phone application that allows users to identify organisms on photos using automatic image recognition (Seek, 2021). Furthermore, taxonomic identification of organisms can be validated by experts within iNaturalist to achieve ‘research grade’ status (see https://www.inaturalist.org/pages/help#quality). The platform has even been used to create a specific project for ‘medically important venomous snakes’ (Genevieve et al., 2018), which now contains over 12,000 georeferenced observations from 285 species by 3440 observers (https://www.inaturalist.org/observations?project_id=10715).
Citizen science platforms could prove valuable in filling sampling gaps (Chandler et al., 2017), especially if contributions from poorly sampled regions can be elicited (Genevieve et al., 2018). Further data can be extracted directly from social media platforms along with coordinates automatically recorded by smartphones (Barve, 2014). However, a suitable system to vet citizen science data rigorously needs to be established before integration into research grade datasets. Some vetting tools may include superior image recognition systems, crowdsourcing of snake identification (Durso et al., 2021), and data pipelines for targeted expert vetting of priority data or areas. These systems could be integrated into the new WHO database mentioned above, which is already planned to include an interactive map viewer of expert vetted snake distributions, species photos and information on antivenoms and antivenom producers, and will provide a ‘one-stop-shop’ for data access and collaboration between researchers, stakeholders, and the general public. It will function as a nexus to continuously update taxonomy and distributions based on literature and occurrence data from a broad range of databases under consultation with an expert panel and contributions from the general public (David Williams, pers. Com.).
As mentioned, simple presence points or area maps of snake distributions are informed by occurrence records, maps in scientific publications, expert knowledge, and subjective interpretations of connectivity between clusters of distribution records. In the age of ‘big data’ (Leonelli, 2014; sourcing, processing and analysis of large datasets using information technology) and high performance computing systems, such bias can be greatly reduced using statistical methods that describe species' habitat suitability, referred to as ecological niche models (ENMs; Sillero, 2011), should sufficient input data exist. A large suite of ENM methods has evolved over the last two decades, many of which are already extensively used in conservation (Guisan and Thuiller, 2005; Guisan et al., 2013; Mizsei et al., 2020) and epidemiology of zoonotic diseases (Escobar and Craft, 2016; Escobar et al., 2013; Murray et al., 2018; Peterson, 2014; Soucy et al., 2018). ENMs use known occurrence localities and environmental conditions to estimate environmental suitability across the study area and predict potentially occupied habitat (Fig. 1). The availability of increasingly fine-scale, gridded geographic data on land use, climate, vegetation, topography, and other landscape features enable prediction of suitable habitats for a species, how suitability varies between grid cells, and when linked to back-casts or future projections of these factors also how it may have changed in the past or will change in the future. Reliable ENMs can often be created with reasonably small data sets (20–50 occurrence records; Stockwell and Peterson, 2002) and for large batches of species using high-performance computing infrastructure (Pintor et al., 2018, 2019). ENMs can help delineate boundaries of suitable habitat around known occurrences objectively, detect habitat patches that are suitable but unsampled (Terribile et al., 2018; Yousefi et al., 2015), determine the degree of habitat connectivity, describe the likelihood of snake encounters as opposed to simple presence or absence (Yañez-Arenas et al., 2018), and generally increase the resolution of distribution maps. In essence they enable description of the area of occupancy (actually occupied habitat patches) within a snakes' extent of occurrence (approximate outline encompassing all occurrences; IUCN, 2020).
ENMs have already been used to predict distributions of venomous species for studies on biogeography, phylogeography, or conservation (Asadi et al., 2019; Barlow et al., 2013; Brito et al., 2008; Burbrink and Guiher, 2015; Di Cola and Chiaraviglio, 2011; Gül; Terribile et al., 2018; Yousefi et al., 2015), and to estimate human risk of exposure to snakebite (Bravo-Vega et al., 2019; Nori et al., 2014; Saupe et al., 2011; Yañez-Arenas et al., 2018; Yañez-Arenas et al., 2014; Yañez-Arenas et al., 2016; Yousefi et al., 2020; Zacarias and Loyola, 2019, Table 1). The most commonly used ENM method amongst the set of studies in Table 1, and probably amongst ENM literature in general, is Maxent. Maxent (i.e., the maximum entropy algorithm; Phillips et al., 2006; Phillips and Dudík, 2008) is a machine learning algorithm that performs well compared to many other methods (Elith et al., 2006), especially when working with presence only datasets, i.e. without ‘true absences’ where the species is known not to occur. Presence only datasets are common, especially for data deficient species, because substantial sampling effort is needed to confirm a species' absence from a location with certainty while confirming its presence only requires one observation (Phillips et al., 2009). Other commonly used methods are boosted regression trees (BRTs; Elith et al., 2006; Elith and Leathwick, 2017; Elith et al., 2008), generalized linear models (GLM; Guisan et al., 2002; McCullagh, 2019), generalized additive models (GAM; Grego, 2006; Guisan et al., 2002; Hastie and Tibshirani, 1987; Hastie and Tibshirani, 1990; Liu, 2008), generalized boosting models (GBM; Ridgeway, 2007), Artificial Neural Networks (ANN; Colasanti, 1991; Lek and Guégan, 1999), random forest models (RF; Breiman, 2001; Evans et al., 2011), Integrated Nested Laplace Approximation (INLA) Bayesian methods for fitting models with spatial random effects (R-INLA; Lindgren and Rue, 2015; Redding et al., 2017), and the genetic algorithm for rule-set production (GARP; Stockwell, 1999). Often several methods are combined into ensemble models to allow uncertainty to be quantified by comparing where models disagree and to compare model performance more generally since novel advances of existing model methods occur frequently (Araújo and New, 2007; Diniz-Filho et al., 2009).
Table 1.
Summary of example studies using ENMs to estimate snake species distributions for a variety of purposes, including epidemiology of snakebite, snakebite risk, and snakebite incidence.
| ENM Method° | Time | Resolution | Species | Geographic Area | Purpose | |
|---|---|---|---|---|---|---|
| Brito et al. (2008) | Maxent | current | ~1 km | Vipera latastei | Southern Europe Northern Africa | Phylogeography |
| Vipera monticola | ||||||
| Di Cola and Chiaraviglio (2011) | GARP | current | ~10 km | Bothrops alternatus | Argentina | Biogeography |
| Bothrops ammodytoides | ||||||
| Bothrops diporus | ||||||
| Lawing and Polly, 2011 | Bioclim | 2100 | 2.5 arc-minutes | 11 rattlesnakes in the genus Crotalus | North America | Biogeography |
| GLM | −6000 | Conservation | ||||
| −21000 | ||||||
| Yañez-Arenas et al. (2016) | Maxent | current | ~20 km | 192 species of venomous snakes | North America | Snakebite Incidence* |
| 2050 | Central America | |||||
| South America | ||||||
| Barlow et al. (2013) | Maxent | −21,000 | 2.5 arc-min | Bitis arietans | Africa | Phylogeography |
| Lyet et al., 2013 | GAM | current | 50 m | Vipera ursinii | France | Ecology |
| Conservation | ||||||
| Yañez-Arenas et al., 2014 | GARP | current | ~1 km | 21 species of venomous snakes | Veracruz, Mexico | Snakebite Incidence* |
| Nori et al. (2014) | Ensemble: | current | ~5 km | Bothrops alternatus | Argentina | Snakebite Risk* |
| Maxent | 2030 | Bothrops ammodytoides | ||||
| GARP | 2080 | Bothrops diporus | ||||
| SVM | Crotalus durissus terrificus | |||||
| Micrurus pyrrhocryptus | ||||||
| Burbrink and Guiher (2014) | Maxent | current | ~1 km | Agkistrodon piscivorus | North America | Phylogeography |
| Agkistrodon contortrix | ||||||
| Agkistrodon conanti | ||||||
| Agkistrodon laticinctus | ||||||
| Yousefi et al. (2015) | Maxent | −21,000 | ~1 km | Montivipera raddei species complex | Iran | Ecology, |
| current | Turkey | Conservation | ||||
| 2070 | Armenia | |||||
| Gül (2015) | Maxent | current | ~1 km | Vipera barani | Turkey | Conservation |
| Mizsei et al. (2016) | Maxent | current | ~1 km | Vipera ursinii | Albania | Conservation |
| Schield et al., 2018 | Maxent | −21000 | 2.5-min | Crotalus scutulatus | Biogeography | |
| Terribile et al. (2018) | Bioclim | current | 0.5° resolution | Micrurus lemniscatus species complex | South America | Conservation |
| ENFA | 2080–2100 | |||||
| Euclidian Distance | ||||||
| FDA | ||||||
| GAM | ||||||
| GLM | ||||||
| Gower Distance | ||||||
| Mahalanobis Distance | ||||||
| MARS | ||||||
| Maxent | ||||||
| ANN | ||||||
| RF | ||||||
| Yañez-Arenas et al. (2018) | Maxent | current | ~1 km | 39 species of venomous snakes | Ecuador | Snakebite Risk* |
| Strickland et al. (2018) | Maxent | current | ~1 km | Crotalus scutulatus | Venom study | |
| Longbottom et al. (2018) | BIOCLIM | current | ~5 km | 278 species of venomous snakes | Global | Snakebite Risk* |
| Asadi et al. (2019) | Ensemble: | current | ~1 km | Gloydius caucasicus | Iran | Phylogeography |
| Maxent | Conservation | |||||
| GLM | ||||||
| GBM | ||||||
| RF | ||||||
| Zacarias et al. (2019) | Maxent | current | ~5 km | Atractaspis bibronii | Mozambique | Snakebite Risk* |
| 2080 | Bitis arietans | Conservation | ||||
| Bitis gabonica | ||||||
| Causus rhombeatus | ||||||
| Dendroaspis angusticeps | ||||||
| Dendroaspis polylepis | ||||||
| Dispholidus typus | ||||||
| Naja annulifera | ||||||
| Naja melanoleuca | ||||||
| Maja mossambica | ||||||
| Naja nigricollis | ||||||
| Thelotornis capensis | ||||||
| Thelotornis usambarics | ||||||
| Bravo-Vega et al. (2019) | Maxent | current | ~1 km | Bothrops asper | Costa Rica | Snakebite Incidence* |
| Lourenço-de-Moraes et al., 2019 | Bioclim, Maxent, ENFA | 2080 | 0.05° | 144 species of snakes including 24 venomous snakes | Brazil, | Conservation |
| Mizsei et al. (2020) | Ensemble | current | Vipera graeca | Greece and Albania | Ecology | |
| GLM | 2020 | Conservation | ||||
| GAM | 2040 | |||||
| ANN | 2060 | |||||
| RF | 2080 | |||||
| Maxent | ||||||
| Lara-Galván et al. (2020) | BIOCLIMBIOCLIM.DISMO | current | ~1 km | Crotalus aquilus | Mexico | Ecology |
| BRT | Crotalus atrox | Conservation | ||||
| CART | Crotalus basiliscus | |||||
| FDA | Crotalus Lepidus | |||||
| GAM GLM GLMNET MARS | Crotalus molossus | |||||
| MAXENTMAXLIKE MDF | Crotalus polystictus | |||||
| RF | Crotalus pricei | |||||
| RPART SVM | Crotalus scutulatus | |||||
| Crotalus willardi. | ||||||
| Yousefi et al. (2020) | Ensemble: | current | ~1 km | Macrovipera lebetina | Iran | Snakebite Risk* |
| Maxent | Echis carinatus | |||||
| GBM | Pseudocerastes persicus | |||||
| GAM | Naja oxiana | |||||
| GLM | ||||||
| RF |
°Maxent: maximum entropy models; GLM: generalized linear models; GBM: generalized boosting models; GAM: generalized additive models; RF: random forest models; GARP: Genetic Algorithm for Rule-set Production; SVM: Support Vector Machine.
*Exposure here refers to how likely human populations are to be exposed to a venomous animal based on its distribution and habitat suitability, while risk involves the exposure and its potential consequence, and incidence is the correlation of predictors with explicitly measured numbers of snakebite.
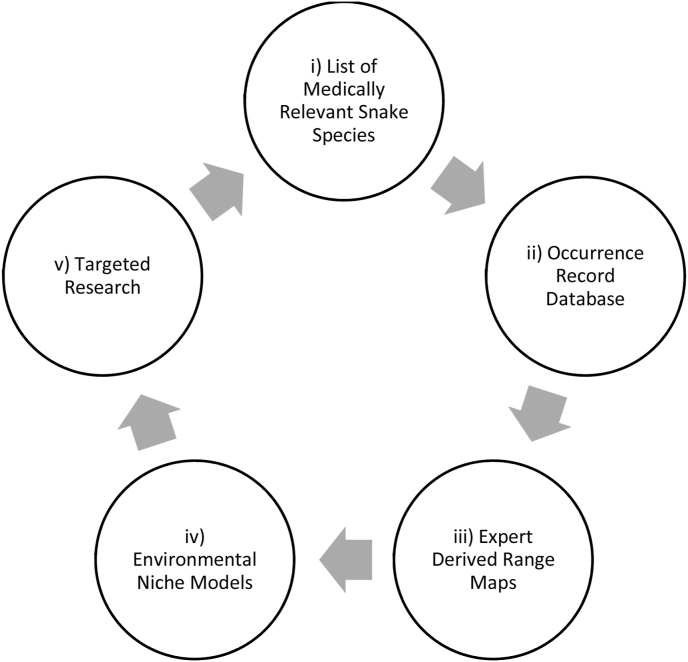
Knowledge of snake distributions is fundamental to understanding where vulnerable human populations are exposed to snakebite, the degree of exposure, and where antivenom for each species is needed. As such, they form the basis for all other aspects of snakebite management and for conservation. Consequently, we recommend a thorough, iterative, globally consistent approach to fill knowledge gaps, where each component is updated regularly and feeds into improvements of the next (Fig. 2). The components are (i) an up-to-date list of medically relevant snakes, (ii) a database of expert vetted occurrence localities for each species, (iii) mapped range estimates based on occurrences, literature, and expert advice, and (iv) ENMs based on known occurrences and high quality, biologically relevant geographic layers of environmental conditions. ENMs ultimately feed into (v) targeted research. The snake master list is updated regularly based on novel taxonomic and epidemiological data. New occurrence data is added from publications, public databases and vetted citizen science data. Range maps are updated under expert advice. ENMs are rerun using new data and environmental layers. Lastly, ENMs can provide information on where additional sampling efforts are needed, or where taxonomy needs revision (e.g. disjunct populations). Efforts to address these knowledge gaps, such as targeted research, then feed back into the master list, the occurrence database and so on. Targeted surveys or elicitation of citizen science efforts in specific, under-sampled areas is required to strategically fill sampling gaps. Note that expert derived range estimates always remain an important part of the process because ENMs only describe habitat suitability but cannot account for other reasons that affect species niche occupation, such as presence of competitor species or inability to reach disjunct patches of suitable habitat. Furthermore, ENMs performance requires validation using expert derived range estimates to account for information that is not available as spatial predictor layers.
Fig. 2.
Proposed components of iterative strategy to improve knowledge on snake species and their distributions.
3. Human-wildlife conflict meets epidemiology
Historically, human-wildlife conflict has been an important issue (Anand and Radhakrishna, 2017; Lamarque et al., 2009; Nyhus, 2016; Treves et al., 2006). The modification of natural habitat for human uses such as farming has led to a myriad of conflicts between humans and wildlife, such as predation of stock by wild predators (Beattie et al., 2020; Hill, 2015; Manral et al., 2016; Messmer, 2000; Western et al.), destruction of crops by herbivores (Kiffner et al., 2021; Mamo et al., 2021; Priston and Underdown, 2009; Siljander et al., 2020), attacks on humans (Jhala et al., 2021; Tarrant et al., 2020; Western et al.), and introduction of zoonotic diseases (Jacob et al., 2020; Jhala et al., 2021; Jones et al., 2013; Tarrant et al., 2020).
Spatial analyses have long been used to study human-wildlife conflict (Carter et al., 2020; Goswami et al., 2015; Kretser et al., 2008; Laliberte and Ripple, 2003; Siljander et al., 2020). For example, Siljander et al., (2020) combined a georeferenced dataset of interviews with statistical geographic analyses over land use maps to understand the geographic patterns of crop raiding by non-human primates in Kenya, enabling appropriate preventative measures by identifying the most vulnerable locations. Similarly, Goswani et al., (2015) used mechanistic modelling to understand the patterns of crop raiding by elephants in India to make management recommendations.
Epidemiology has also frequently used spatial analyses to estimate the spread of diseases (Peterson, 2014; Santos-Vega et al., 2016). Spatial Epidemiology has blossomed with the advent of big data, geostatistical methods and increased computing power, resulting in a movement termed precision public health (the combination of high-resolution health data with environmental and socioeconomic predictors to produce fine-scale estimates of disease risk; Desmond-Hellmann, 2016). For diseases spread directly amongst primary hosts, without the need of a vector or reservoir, simple mathematical models describing host interaction frequencies and disease transmission rates are usually sufficient to estimate disease spread (Grassly and Fraser, 2008). However, it has recently been emphasized that disease transmission risk has an important but often neglected ecological component dependent on the distribution, habitat requirement, and ‘population’ dynamics of both the pathogen and host species (Peterson, 2014).
In the special case of zoonotic diseases, epidemiological studies have the added challenge of mapping several biotic components of the disease transmission: these include wildlife that functions as disease reservoirs and, in some cases, disease vectors that spread the infectious agent between reservoirs and primary hosts (for example mosquitos). As such, zoonotic and vector borne diseases present an intersection between human-wildlife conflict and traditional epidemiology (Reisen, 2010). In cases where data on disease itself is sparse, as is common for NTDs and emerging infectious diseases, vector and host distribution often serve as a useful metric of risk to guide preventative measures (Campbell et al., 2015; Ferro et al., 2015; Mylne et al., 2015; Peterson, 2014) as the pathogens spread depends greatly on the population dynamics and abundance of vector and host (Lloyd-Smith et al., 2005). Similarly, the pathogen's habitat requirements are determined by the internal conditions of the vector and host, therefore, areas of disease risk can be seen as the intersection of vector, host, and pathogen distributions (Reisen, 2010) or species richness resulting from distribution overlap (Ferro et al., 2015). There is a multitude of studies that illustrate how spatial analyses can disentangle relevant epidemiological patterns in zoonotic diseases and NTDs (Hamm et al., 2015; Luz et al., 2010; Marshall, 1991), often by interpolation of important spatial features of disease dynamics from limited source data to unsampled locations.
While snakebite is similar to zoonotic diseases in some respects, such as the involvement of both a human victim and a wildlife agent inflicting the disease, it has unique attributes compared to such diseases. In many ways, snakebite has more in common with traditional human-wildlife conflicts that involve physical harm inflicted on humans, not least because it involves conservation concerns of the ‘agent’ (Pandey et al., 2016). However, while not caused by a pathogen, snakebite envenoming is more analogous to a disease than a physical injury because it involves complicated and prolonged physiological and immunological effects and treatments (Gutiérrez et al., 2011; Ogawa et al., 1996; Russell, 1988) and, consequently, has rightfully been elevated to NTD status (WHO, 2017). Snakebite risk can also be viewed as the result of overlaps in snake and human distributions, modified by patterns in their abundance, activity, and population dynamics, similar to vector borne diseases. Consequently, the same tools that have been used to disentangle spatial patterns in human-wildlife conflicts and vector borne diseases can be applied to snakebite research. This path has begun to receive attention, but progress is currently limited by sparse data on snake distributions, spatial ecology, general biology, and snakebite incidence (see following sections).
4. Spatial patterns in diversity, abundance, activity, and population dynamics of snakes
How humans interact with snakes depends on snake distributions, and how humans and snakes overlap in their use of space and time within those distributions (Goldstein et al., 2021). This, in turn, depends on snake abundance, activity patterns, and population dynamics. Unfortunately, all three of these attributes of snake biology are understudied.
The abundance of any species varies across their distribution (Brown, 1984), depending on how it uses the available space (i.e. behavioral requirements such as preferred foraging habitats; Blouin-Demers and Weatherhead, 2001) and how favorable different habitats are to population growth (i.e. physiological requirements, such as temperature regime; Medina-Barrios et al., 2019). Studies quantifying the variation in abundance of snakes across their distribution are sparse (Bravo-Vega et al., 2019), costly, and time consuming. ENMs aim to estimate species' realized ecological niche (as opposed to the ‘occupied niche’ which represents the subset of conditions that are historically and geographically accessible; Sillero, 2011) and, therefore, provide estimates of habitat suitability. Theoretically, higher habitat suitability should coincide with higher abundance, as long as all relevant environmental features that influence a species' behavioral and physiological requirements are included as predictors (Ehrlén and Morris, 2015; Jiménez-Valverde et al., 2021; VanDerWal et al., 2009; Weber et al., 2017), although this trend is contentious (Dallas et al., 2017; Dallas and Hastings, 2018). Consequently, habitat suitability derived from ENMs is often used as a proxy for abundance, or at least of upper limits of potential abundance, since unknown factors that are not included in models (e.g. presence of predators, competitors or unknown environmental variables) may further limit abundance (Braz et al., 2020; Jiménez-Valverde et al., 2021; Muñoz et al., 2015; VanDerWal et al., 2009; Weber et al., 2017). Additionally, the observed relationship between habitat suitability and abundance may not be linear but asymptotical (VanDerWal et al., 2009) as abundance approaches carrying capacity and may be weakened due to dispersal amongst neighboring cells with different suitability, especially when resolutions are high compared to dispersal ability (Macartney et al., 1988). Nevertheless, correlations of ENM derived habitat suitability with upper limits of abundance have been observed (Braz et al., 2020; Jiménez-Valverde et al., 2021; VanDerWal et al., 2009; Weber et al., 2017). In fact, snakes' habitat suitability or metrics based on it (such as distance from the ‘niche centroid’; Yañez-Arenas et al., 2016) have been used as proxies of snake abundance and snakebite risk and have even been shown to correlate with snakebite incidence (Yañez-Arenas et al., 2016). As such, we encourage studies that further test the ability of ENMs to accurately predict abundance across different species and identify how ENMs predictive ability of abundance can be improved.
Even in areas of high snake abundance, humans are only exposed to snakebite risk if snakes are actually active at the same time as people, and there is overlap within the same geographic space (Goldstein et al., 2021). Reptile activity and microhabitat selection varies with season (Ediriweera et al., 2018; Lindström et al., 2015; Madsen and Shine, 1996) time of day (Ealy et al., 2004), and ambient abiotic conditions (Pintor et al., 2016), as do human activity patterns (Goldstein et al., 2021). These temporal patterns in activity are usually a direct result of (i) patterns in abiotic conditions (higher activity at warmer temperatures or after rain; Angarita-Gerlein et al., 2017; Karabuva et al., 2016) and (ii) biological factors, such as breeding seasons or increases in prey abundance (Ediriweera et al., 2018). The latter, in turn, are triggered by changes in abiotic conditions (Licht, 1972). Because most changes in activity patterns are ultimately influenced by abiotic conditions, they can be modelled using average monthly conditions (climate oscillations; for determining typical seasonal patterns) and daily historic weather data (weather anomalies; for determining weather related deviations from seasonal averages; (Ediriweera et al., 2018). Fine-scale spatio-temporal climate and weather data has become available for variables such as temperature and precipitation (Fick and Hijmans, 2017; Funk et al., 2015), but also for resulting changes in habitat attributes (e.g. 10-daily 300 m resolution layers of global fraction photosynthetic active radiation; Fuster et al., 2020). Historical weather data has already been used to model spatio-temporal variation in habitat use by nomadic animals (Reside et al., 2010) and to disentangle the effects of seasonal climate patterns versus weather anomalies on temporal variation in snakebite incidence in Sri Lanka (Ediriweera et al., 2018). Dynamic models of how snake activity and abundance vary across time and space could prove useful as forecasting tools to predict when people may experience elevated risk of encountering snakes and which species are encountered more at different times of year. Such forecasts could allow health centers to prepare for increased numbers of snakebite patients or to warn the public to take additional precautions to avoid snakebite. Together with information on circadian rhythms of snakes, very fine-scale (i.e. 10 m × 10 m) spatio-temporal models of snakebite risk could be created (Goldstein et al., 2021).
Spatio-temporal patterns in snake presence, abundance and activity lead to complex patterns in snake diversity, which also affect snakebite risk. Although some snakes are more prone to bite than others because they enter human dwellings, are harder to see, or are more aggressive (Goldstein et al., 2021), the overall degree of human exposure results from the cumulative exposure to all species present in an area. Consequently, patterns in snake diversity are a crucial aspect of variation in snakebite risk. It has been proposed that snakebite risk can be estimated using the cumulative snake species richness weighted by each species' propensity to inflict bites (e.g. the known fraction of bites caused by each species in a country or district; Yañez-Arenas et al., 2016; Zacarias and Loyola, 2019) but further research needs to establish how different species’ presence, habitat suitability, and biting propensity interact to lead to differences in overall snakebite risk.
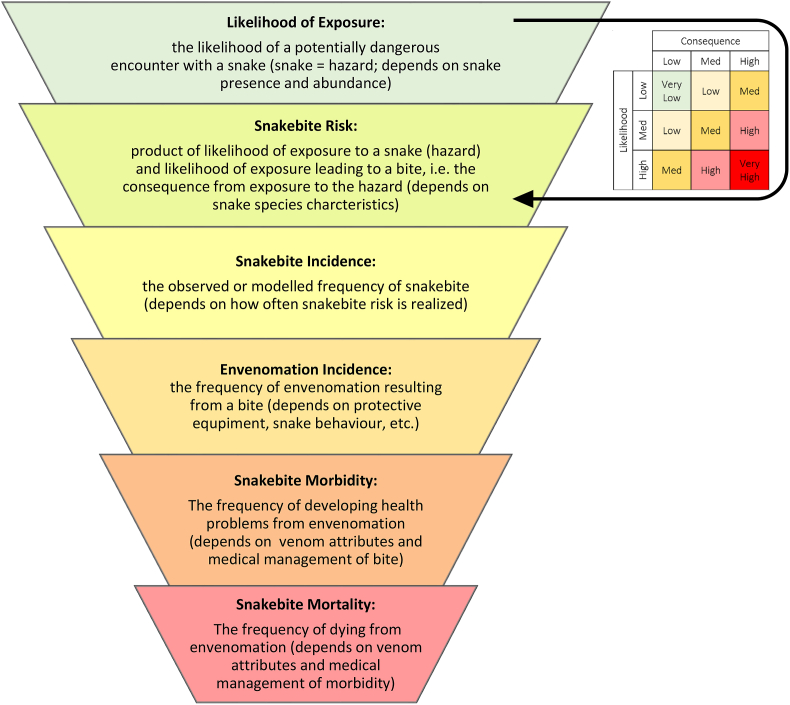
It is also noteworthy that cumulative weighted snake species richness is a measure of snakebite risk, i.e. the product of the likelihood of exposure to a snake (snake presence and abundance = exposure to the hazard) and the likelihood of an encounter leading to a bite (e.g. snake's propensity to bite = potential consequence of exposure to the hazard; Fig. 3). The terms ‘snakebite risk’ and ‘snakebite incidence’ are often used interchangeably and often also applied to mere snake exposure (WHO, 2010b; Yañez-Arenas et al., 2018). We suggest that snakebite risk is henceforth used to describe the theoretical probability of encountering, and being bitten by a snake, while incidence is the realized, observed or modelled snakebite frequency and depends on additional factors such as human activities, demography, population density, and protective equipment, amongst others, i.e. how often snakebite risk is realized (Fig. 3). In lay terms, snakebite risk is the likelihood that one could encounter a snake and be bitten by it in a given area at a given time. Snakebite incidence is the frequency at which these encounters lead to actual bites based on how many people are in the area, their activity patterns, their awareness of the risk, and how they manage the encounter. Snakebite risk is unlikely to change if snakes are conserved successfully because it relates to features intrinsic to snakes present in the area, while snakebite incidence can be reduced with adequate education and management (Ediriweera et al., 2018). Following this, snakebite envenoming, snakebite related morbidity and mortality are influenced by snakebite incidence. The former depends on protective equipment, the snake's agitation, and its behavioral propensity to inflict wet bites. The latter two depend on how well snakebite is managed from a medical perspective.
Fig. 3.
Diagram describing the dependence of snakebite mortality and morbidity on snakebite and envenomation incidence, and risk (the product of likelihood of exposure and consequence of exposure). Snakebite risk is intrinsic to the nature of the dangerous herpetofauna in an area, incidence is how often the risk is realized, and snakebite morbidity/mortality further depend on snakebite management practice.
Note that modification of human activities can alleviate snakebite risk. Some may, therefore, choose to include them in the risk definition. However, determining the effect human activities in an area on risk usually requires knowledge of actual snakebite numbers and is, consequently, hard to separate from observed incidence. In the literature, human activities are almost always included in analyses of observed incidence, not theoretical risk (which can be mapped without knowledge of actual snakebite numbers). In theory, however, the expected rather than observed effect of different activities on snakebite risk could be mapped and, in such cases, it may be considered as a modifying factor of risk, rather than of incidence (e.g. the theoretical risk of snakebite for a farmer using machinery versus manual labor).
Several recent studies have estimated geographic variation in snakebite risk using modelled snake diversity (i.e. cumulative presence-absence maps) or some measure of cumulative habitat suitability (as a proxy for cumulative abundance; Yañez-Arenas et al., 2018 [Ecuador]; Yousefi et al., 2020 [Iran]; Zacarias and Loyola, 2019 [Mozambique]). Some have even confirmed a correlation between snakebite risk and snakebite incidence (Yañez-Arenas et al., 2016 [Americas]; Yañez-Arenas et al., 2014 [Mexico]). It would be useful to expand snakebite risk maps globally, to estimate spatial variation and seasonal and weather based fluctuations in snakebite risk, and to perform rigorous ground-truthing of these modelling approaches’ ability to estimate spatio-temporal variation in snake activity, abundance, and diversity.
5. The missing link: how do humans & snakes interact to create spatio-temporal patterns in snakebite incidence
Similar to how the frequency and type of human-snake interactions depend on snake abundance, activity, and population dynamics, they also depend on human population density, lifestyle, and demographics. Many studies worldwide have documented demographic patterns with respect to snakebite epidemiology (Ediriweera et al., 2016). Across most countries, young males in rural communities, agricultural workers, and members of lower socio-economic and less well-educated groups are disproportionately affected (Dehghani et al., 2014; Harrison and Gutiérrez, 2016; Harrison et al., 2009; Suraweera et al., 2020). Patterns of spatial variation in snakebite incidence usually follow these general epidemiological patterns: at a global scale, snakebite incidence varies greatly, with hotspots in regions with rural subsistence farming such as South Asia, tropical sub-Saharan Africa and Latin America (Kasturiratne et al., 2008). At intermediate scales, snakebite incidence or mortality has been documented nationally for countries in Africa, Europe, the Americas, and South Asia (Chippaux, 1998, 2011, 2012, 2017; Halilu et al., 2019; Suraweera et al., 2020). At a fine scale, for much of the Americas, and some of South Asia and Africa, some data exists at district or municipality level (Bravo-Vega et al., 2019; Chaves et al., 2015; Chippaux, 2017; Ediriweera et al., 2016; Hansson et al., 2010, 2013; León-Núñez et al., 2020; Mohapatra et al., 2011; Molesworth et al., 2003; Yañez-Arenas et al., 2014, 2016).
Potential drivers of spatial snakebite variation at intermediate scales have been quantified to some extent, using anything from simple statistics such as t-tests (Chippaux, 2017; León-Núñez et al., 2020) to more elaborate statistical models such as generalized additive models (GAM; Ediriweera et al., 2016), geostatistical binomial logistic models (Ediriweera et al., 2018), spatial Poisson models (Suraweera et al., 2020) or bottom-up agent-based models (Goldstein et al., 2021); Table 2). Again, hotspots tend to occur in rural, agricultural, and poor areas (Chaves et al., 2015; Ediriweera et al., 2016; Hansson et al., 2010, 2013; Leynaud and Reati, 2009; Schneider et al., 2021; Suraweera et al., 2020), and more bites occur in young to middle aged males or in regions with a higher male population percentage (Chippaux, 2017; Ediriweera et al., 2016; Hansson et al., 2010; León-Núñez et al., 2020; Mohapatra et al., 2011; Suraweera et al., 2020). Relationships between spatial snakebite variation and human population density are more complex: usually snakebites increase with human population density in rural areas but drop off at higher densities associated with urbanization (Chippaux, 2017; Ediriweera et al., 2016). As expected, snakebite incidence also correlates with measures of presence, activity, abundance, or diversity of snakes (Bravo-Vega et al., 2019; Goldstein et al., 2021; Hansson et al., 2013; León-Núñez et al., 2020; Schneider et al., 2021; Suraweera et al., 2020; Yañez-Arenas et al., 2014, 2016) or with variables that affect snake activity. Often snakebite incidence increases during certain seasons when snakes and farmers are both more active such as in rainy or harvest seasons (Chippaux, 2017; Ediriweera et al., 2018; Goldstein et al., 2021; Hansson et al., 2010; Mohapatra et al., 2011; Molesworth et al., 2003; Patiño-Barbosa et al., 2019; Suraweera et al., 2020), during flooding events (Ochoa et al., 2020), or at higher temperatures, lower altitudes, and higher precipitation (Angarita-Gerlein et al., 2017; Chaves et al., 2015; Chippaux, 2017; Ediriweera et al., 2018; Ediriweera et al., 2016; Goldstein et al., 2021; Hansson et al., 2013; Schneider et al., 2021; Suraweera et al., 2020, Table 2).
Table 2.
Summary of key studies on spatial variation in snakebite incidence or mortality, ranging from simple descriptive studies to fine-scale predictions.
| Type | Measure | Area | Resolution | Method | Important Predictors | |
|---|---|---|---|---|---|---|
| Studies describing broad scale spatial patterns and hotspots in snakebite incidence | ||||||
| Swaroop (1954) | Spatial* | Mortality | Global | Source data: | NA | NA |
| Temporal* | Country | |||||
| Predictions: | ||||||
| NA | ||||||
| Chippaux (1998) | Spatial | incidence | Global | Source data: | NA | NA |
| Country | ||||||
| Predictions: | ||||||
| Snakebite Regions | ||||||
| Kasturiratne et al. (2008) | Spatial | Incidence | Global | Source data: | NA | NA |
| Mortality | Countries | |||||
| Predictions: global burden region | ||||||
| Studies using simple statistics, epidemiology, and coarse scale spatial predictors to describe spatial variation in snakebite incidence | ||||||
| Molesworth (2003) | Spatial | Incidence | West Africa (Ghana & Nigeria) | Source data: | LogR | NDVI↑ |
| Temporal | 29 health facilities Predictions: ~15 km grid | Season (Rainy season) | ||||
| Leynaud and Reati (2009) | Spatial | Incidence | Cordoba, Argentina | Source data: | Spatial smoothing model | Location in departments with high percentage of persistence farming |
| Department | Species identity | |||||
| Predictions: | ||||||
| department | ||||||
| Mohapatra et al. (2011) | Spatial | Mortality | India | Source data: | LogR | Male/Female |
| Temporal | ~7000 small areas | Religion (Hindu↑) | ||||
| Individual | Predictions: states | Occupation (Agricultural worker↑) | ||||
| Season (Monsoon↑) | ||||||
| State (high prevalence states↑) | ||||||
| Age (15–29↑) | ||||||
| Chippaux, 2017** | Spatial | Incidence | Americas | Source Data: | t-test, Pearson Correlation, Chi Squared, Mann-Whitney Test | Altitude↓ |
| Temporal | mortality | Province | Male/Female | |||
| Individual | Predictions: | Age (young to middle aged↑) | ||||
| Province | Climate Zone | |||||
| Season (Rainy or Summer↑) | ||||||
| Population density↑↓ | ||||||
| Year↑↓ | ||||||
| Angarita-Gerlein et al. (2017) | Spatial | incidence | Colombia | Source Data: | Cross-correlation analysis | Precipitation |
| Temporal | Municipality | Municipality Identity | ||||
| Predictions: | ||||||
| Municipality | ||||||
| Riascos et al., 2019 | Spatial | Incidence | Coffee Triangle Region, Colombia | Source data: Municipality | NA | Year |
| Temporal | Predictions: | Season | ||||
| NA | ||||||
| León-Núñez et al. (2020) | Spatial | Incidence | Colombia | Source data: | t-test, Pearson Correlation, Chi Squared, Mann-Whitney Test | Male/Female |
| Individual | Department | Urban/Rural | ||||
| Predictions: Department | Ethnicity (Afro-Colombian & Indigenous↑) | |||||
| Age (28–35↑) | ||||||
| Region (Amazonia & Orinoquia↑) | ||||||
| Species identity | ||||||
| Year↑ | ||||||
| Studies using relatively novel fine scale source data, advanced statistical models, and improved resolution | ||||||
| Hansson et al. (2010) | Spatial | Incidence | Nicaragua | Source data: municipality | Poisson regression | Season (Rainy Season↑) |
| Temporal | Predictions: | Environmental Region (altitude, precipitation, geographic clustering; Wet Lowlands↑) | ||||
| Individual | municipality | Rural population percentage↑ | ||||
| Male population percentage↑ | ||||||
| Young population percentage | ||||||
| Underreporting index↑ | ||||||
| Hansson et al. (2013) | Spatial | Incidence | Costa Rica | Source data: district | Bayesian Poisson regression | altitude↓ |
| Predictions: | precipitation↑ | |||||
| district | length of dry season↓ | |||||
| rural population percentage↑ | ||||||
| population percentage near large forests↑ | ||||||
| Snake habitat suitability↑ | ||||||
| Chaves et al. (2015) | Spatial | incidence | Costa Rica | Source data: County | geographically weighted regression | Weather & Climate Oscillations |
| Temporal | Predictions: | Temperature↑ | ||||
| County | Precipitation | |||||
| Poverty Indicators (Poverty gap | ||||||
| index and percentage of destitute housing)↑ | ||||||
| Altitude↓ | ||||||
| Yañez-Arenas et al. (2016) | Spatial | Incidence | Americas | Source data: | GLM | Cumulative MRS presence & abundance index (SRI_2) ↑ |
| Provinces Predictions: ~20 km grid | ||||||
| Yañez-Arenas et al., 2014 | spatial | Incidence | Veracruz, Mexico | Source data: Municipality | GAM | 2 MRS species' abundance estimate↑ |
| Predictions: Municipality | Index of marginalization↑ | |||||
| Suraweera et al. (2020) | Spatial | Mortality | India | Source data: | Spatial Poisson model | Age group (30–69↑) |
| Temporal | Incidence (inferred) | ~7000 small areas Predictions: ~50 km grid | Male/Female | |||
| Individual | Season (Monsoon↑) | |||||
| Elevation to 400m↓ | ||||||
| Urban/Rural | ||||||
| Poverty (rural female illiteracy)↑ | ||||||
| Monthly mean temperature to 20 °C↑ | ||||||
| Year↓ | ||||||
| Species identity | ||||||
| Schneider et al. (2021) | Spatial | Incidence | Brazil | Source data: | Negative binomial regression model | Major habitat type (Tropical↑) |
| Municipality | Temperature↑ | |||||
| Predictions: | Precipitation↑ | |||||
| Municipality | Elevation↑ | |||||
| Urbanization percentage↓ | ||||||
| Venomous snake richness | ||||||
| Forest loss↑ | ||||||
| GDP per capita↓ | ||||||
| Studies resulting in fine scale predictions of snakebite incidence | ||||||
| Ediriweera et al., (2016) | Spatial | Sri Lanka | Source Data: household clusters in smallest administrative divisions | GLM GAM Geostatistical binomial logistic Log-linear models |
Male/Female Age (middle aged↑) Time of day (evening↑) Occupation (farm labourer↑) Education↓ Monthly income↓ Population density↓ Elevation Occupation distribution Climatic zone Season humidity weather abnormalities↓ |
|
| Ediriweera et al. (2018) | Temporal | Predictions: | ||||
| Ediriweera et al., 2019 | Individual | 1 km | ||||
| Bravo-Vega et al. (2019) | Spatial | incidence | Costa Rica | Source Data: District Predictions: 1 km |
Linear regression | Encounter frequency of Bothrops asper Human population density |
| Goldstein et al. (2021) | Spatial temporal |
Incidence | Sri Lanka | Source data: 10m-2km Predictions:2 km study squares |
Bottom-up Agent based modelling | Snake-famer activity overlap patterns based on: Monthly precipitation Number of rainy days Farmer type Land type Daily farmer activity time↑ Population percentage farmers↑ Snake activity season↑ Circadian snake activity time↑ Snake aggressiveness↑ Snake land type association↑ Snake abundance estimate↑ |
GAM = generalized additive models; GLM = generalized linear models; LogR = Logistic regression; SRI = ‘snakebite risk index’; NDVI = normalized difference vegetation index.
↑ = positive correlation; ↓ = negative correlation; no arrow = complex correlation pattern; bold text = significant categorical predictor.
*Information given in written form such as tables but could be analysed spatially and/or temporally.
**small scale studies already summarized in this review are not listed again separately in the table.
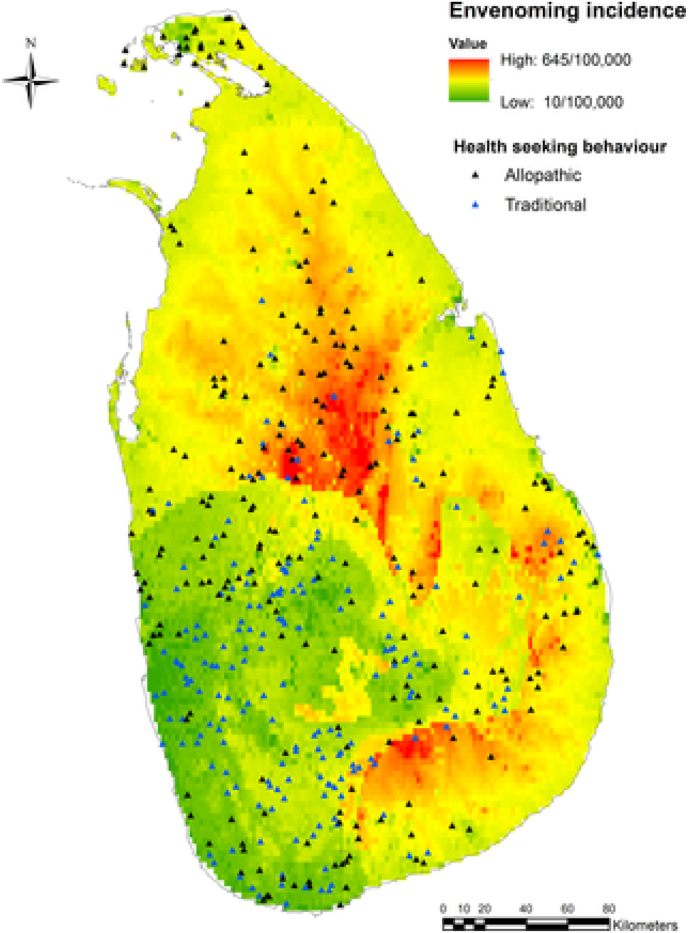
While all these studies have made tremendous contributions to our understanding of spatial snakebite variation, most have not analysed it at spatial resolutions sufficient for on-ground management. The first generation of studies on spatial snakebite variation mostly focused on broad patterns and identified global hotspot regions or inter-country variation (Chippaux, 1998; Kasturiratne et al., 2008; Swaroop and Grab, 1954). Such studies enable estimates of snakebite numbers from incomplete reporting data and help identify areas where intervention or further research is needed. The next suite of studies incorporated simple tests of variables that explain spatial snakebite variation at country, district, or municipality level in combination with epidemiological data on individual risk and temporal patterns (Chippaux, 2017; León-Núñez et al., 2020; Leynaud and Reati, 2009). Most of these made use of the increasingly fine-scale data on snakebite numbers that became available across much of the Americas, India and Sri Lanka relatively recently due to changes in reporting requirements or costly efforts in one-time surveys (Chippaux, 2017; Ediriweera et al., 2016) or novel health surveys (Ediriweera et al., 2016; Mohapatra et al., 2011). These advances led to more complex models within these countries utilizing sophisticated methods such as generalized linear models (GLM), generalized additive models (GAM) and a variety of other frequentist and Bayesian geostatistical regression approaches, incorporating an ever-increasing suite of gridded spatial data on demography, natural environment, climate, weather, and topography (Table 2). Several have also included measures of snake distributions and abundance as predictors of spatial snakebite variation for the first time (Hansson et al., 2013; Yañez-Arenas et al., 2016). However, such studies are currently restricted to areas with better snake or snakebite data, such as the Americas, India and Sri Lanka. Furthermore, models of spatial snakebite variation at sufficiently fine-scale resolutions for on-ground management and redistribution of health care resources (i.e. a resolution of ~5 km or lower) are still sparse. The few notable exceptions are recent work in Sri Lanka (Ediriweera et al., 2016, 2018, 2019; Goldstein et al., 2021) and Costa Rica (Bravo-Vega et al., 2019). Ediriweera et al., (2016) predicted patterns in spatial snakebite variation at 1 km resolution, along with describing health seeking behaviour patterns (Fig. 4), as well as temporal (Ediriweera et al., 2018) and individual level snakebite incidence variation (Ediriweera et al., 2019). Goldstein et al., (2021) further investigated how annual and daily activity patterns of farmers and snakes overlap to cause spatio-temporal fluctuations in snakebite using a bottom-up, agent-based modelling approach at 10 m resolution. These approaches will likely lead to improvements in local snakebite management in Sri Lanka, where snakebite burden is amongst the highest in the World, and region specific antivenoms are lacking (Kasturiratne et al., 2008, 2017; Keyler et al., 2013). In Costa Rica, Bravo-Vega et al., (2019) used a mathematical approach to describe the likelihood of snakebite based on the encounter frequency of humans with the most dangerous snake species in the area and predicted spatial snakebite variation at a 1 km resolution. This approach is more akin to traditional epidemiology, where infection rates depend on transmission rates and on host-vector interaction frequencies (Peterson, 2014). This research adds to previous studies describing spatial snakebite variation in Costa Rica using the same district level source data but notably downscaled predictions to a finer resolution, and is a promising example for many other countries for which district-level data also exist (Chippaux, 2017; Hansson et al., 2013). It also highlights snakebite as an intersection between epidemiology, ecology, and conservation, and the need to consider transdisciplinary approaches. Lastly, promising models of other human-wildlife conflicts have been created using machine learning algorithms at fine spatial scales (Sharma et al., 2020). Broader application of these existing, successful approaches or integration of benefits from each of them into a more complex human-snake conflict framework requires exploration.
Fig. 4.
Health seeking behaviour pattern versus envenoming incidence in Sri Lanka adapted from Ediriweera et al., 2016; 2017. Individual cases are mapped on an envenoming bite incidence map of Sri Lanka. Black triangles show modern medical treatment seeking behaviour, blue triangles show traditional medical treatment seeking behaviour.
In general, effective on-ground management of snakebite requires relatively fine-scale spatio-temporal models of spatial snakebite variation, along with identification of demographic groups that are at particular risk in any given area (i.e. vulnerable human populations). Model resolution needs to be appropriate to the problem in hand, appropriate under consideration of computational limitations, and reasonable considering currently available baseline data (Williams et al., 2012). If the resolution is too coarse (e.g. 50 km), the model cannot accurately inform management actions at a relevant scale. If it is too fine, it increases computational demand without adding any useful additional information. For example, both snakes and humans can easily travel a few kilometres per day and patterns at resolutions finer than this will be diluted by frequent dispersal from neighboring cells. For country-wide snakebite management, a 1 km resolution is likely sufficient to accurately describe relevant landscape and population features that influence human and snake population dynamics and movement. However, some purposes, such as targeted provision of personal protective equipment amongst different farmers in a village might benefit from extremely fine scale predictions (a few meters) of risk and incidence. The scale of analyses needs to be finely tuned to match the planned application. At the appropriate resolution, incidence maps could be used to establish snakebite management centers, direct antivenom to necessary health centers, plan targeted community education, distribute protective equipment to at-risk groups (Ediriweera et al., 2016), estimate snakebite numbers in any given area, inform manufacturers of antivenom demands, and determine which snake species or populations should be catered for during antivenom production for that area. However, fine-scale models are often difficult to construct due to the limited resolution of source data, which is often recorded at second or third administrative country subnational level. The amount of work required to make the fitting of fine-scale models possible varies regionally and nationally but generally demands better, standardized, spatially referenced reporting systems for snakebite.
For example, snakebite is a reportable disease across much of Latin America as of 2000 (Chippaux, 2017); however, enforcement is difficult and many victims still seek traditional healers instead of health centers (Ediriweera et al., 2016). The situation in Africa is much worse: few countries have official reporting systems (e.g. the Kenyan Wildlife Service) or representative household surveys (Cameroon (Alcoba et al., 2021); and a very large proportion of victims attend traditional healers instead of health centers (Newman et al., 1997). Across South and South-East Asia, India and Sri Lanka have high quality data collected either once-off or even consistently across years, and at a useful spatial scale through standardized household surveys (Ediriweera et al., 2016; Mohapatra et al., 2011). Similarly, Nepal has recently begun representative surveys (Alcoba et al., 2021). However, such surveys are effort-intensive and costly - most countries in the region have limited information and research relies on individual hospital records to fill knowledge gaps (Kasturiratne et al., 2008). The latter usually only cover a small proportion of hospitals and victims (Fox et al., 2006) and are not spatially representative (Kasturiratne et al., 2008). Recently, progress has been made to develop appropriate survey methodologies to assess country-wide spatial snakebite variation, and these methodologies have already been used across two countries in South Asia (Nepal) and Africa (Cameroon; Alcoba et al., 2021). Funding and infrastructure to carry out such surveys is limited in many developing countries (Kasturiratne et al., 2008). Ideally, data from surveys, hospital admissions and health authority reporting systems would directly feed into a central global database managed by WHO; however, until better reporting systems are established, several other region-specific steps could improve our understanding of spatial snakebite variation.
Across the Americas, existing information on snakebite at district or municipality level could be combined with finer-scale spatial data to downscale predictions. In a nutshell, spatial snakebite variation could first be predicted at a district scale using variables that are also available at a finer scale (e.g. temperature averaged per district vs. temperature per 1 km grid cell). Observed relationships could then be ground-truthed in selected areas where finer-scale spatial snakebite variation data exists and, if broad scale relationships hold true at finer scales, predictions could be applied more broadly to high-resolution gridded landscapes.
In South Asia, some countries have used analyses of representative household clusters to create predictions of spatial snakebite variation (Ediriweera et al., 2016; Suraweera et al., 2020). Since other countries in South Asia are already starting to implement similar multi-cluster random survey designs (Nepal; Alcoba et al., 2021), efforts could be further expanded to surrounding countries and incidence mapping methods from India and Sri Lanka could be applied to create a uniform methodology across the region. More complete data needs to be collected for most of South-East Asian spatial snakebite variation to facilitate this approach.
Similarly, in Africa new modelling protocols could be developed in countries with existing reporting systems. Results could then be extrapolated to surrounding countries with a similar range of cultural, demographic, and environmental conditions and similar snake species composition. For example, Kenya has a comprehensive country-wide dataset on snakebite incidence from a human-wildlife conflict compensation scheme (Long et al., 2020), which could be used to model spatial snakebite variation and apply results preliminarily to the rest of Eastern sub-Saharan Africa. Nevertheless, sub-Saharan Africa is culturally diverse and overall particularly data-poor in this respect despite being a hotspot for snakebite. There is an urgent need for further data collection in poorly surveyed regions with high snake diversity and political instability, such as throughout the notoriously data poor Congo Basin.
The lack of data on snakebite numbers stands in stark contrast to the enormous amount of other spatial information that is becoming available at finer and finer scales. Much of the demographic, climatic, topographic, and land cover data needed for spatial snakebite variation models exists at an extremely fine-scale across most of the globe, sometimes at resolutions down to 10m (Goldstein et al., 2021). WorldPop (Tatem, 2017; WorldPop, 2021) has 100m resolution data on human population density, births, age and sex structures, pregnancies, and many other demographic factors for most countries. Global climate data exists at 1 km resolution (Fick and Hijmans, 2017; WorldClim, 2021). The European Space Agency has global data on land cover classes and vegetation characteristics at 300m resolution (ESA, 2017; Fuster et al., 2020). The list of high-quality spatial datasets is long. Considering that 1 km would likely be an effective resolution for spatial snakebite variation models, research on the topic lags behind current GIS and computing capacities. Improved data collection on spatial snakebite variation is the single most urgent step that would allow us to catch up on this lag, followed by snake occurrences and abundance data.
We have come a long way in understanding spatial snakebite variation around the world and within countries but need to make substantial improvements in data collection, model resolution, global consistency of modelling approaches and synchronization of data streams and methodologies.
6. Vulnerable human populations and access to life-saving treatments
Envenomation by a snakebite is a medical emergency that requires rapid access to life-saving treatments (antivenom, respiratory support). Delay to treatment has been shown to increase likelihood of complications and death (e.g. da Silva Souza et al., 2018; Iliyasu et al., 2015). While the causes of these delays can be numerous (Banes et al., 2021) the time taken to reach the treatment facility from the patient household (or biting site) is critical and has been shown to greatly impact health outcomes (Habib and Abubakar, 2011). Unfortunately, health care access is particularly poor in developing countries, where snakebite is most common, and varies substantially across and within countries and amongst social classes. Identifying vulnerable populations from both a demographic and spatial perspective is an essential basis for adequate distribution of resources. It has been long recognized that modelling physical accessibility to healthcare can be instrumental for understanding the population coverage of a given health service, identifying vulnerable populations, and optimizing health resource allocation. Ways of modelling access to healthcare are numerous and can differ greatly in terms of the required spatio-temporal data (Delamater et al., 2012; Neutens, 2015; Paez et al., 2019). In low- and middle-income countries where patients must often use a combination of types of transport, and often walk to reach care, modelling approaches based on least-cost path are particularly well suited (Ray and Ebener, 2008). These approaches typically make use of local travelling constraints (e.g., terrain, rivers, barriers to movement) and infrastructures (e.g. road network), associated with the care-seeking behavior (modes and speeds of transport) of the target population, to output a raster of travel time to the nearest health service. Applications of least-cost methods have been done notably to optimize access to emergency obstetric and neonatal care (Chen et al., 2017; Ebener et al., 2019; Kim et al., 2020), to optimize deployment of community health workers (Oliphant et al., pre-print), to assess access to vaccination centers (Joseph et al., 2020) and intensive care units (Barasa et al., 2020), and to model access to emergency services (Ahmed et al., 2019).
Once a travel time model is available, its overlay with the spatial distribution of the target population can inform about population coverage and the location of populations distant from the needed treatments. Combining travel time with additional spatial criteria (e.g. health system metrics, socio-economic characteristics of the population, disease burden) can enable the modelling of vulnerable populations. To model hotspots of population vulnerable to snakebite envenoming at global scale, Longbottom et al. (2018) combined range maps for medically important venomous snake species, travel time to urban centers (as a proxy for geographic access to care), health care quality index (as a proxy metric for severe snakebite-related outcomes), and antivenom availability. However, improving access to snakebite treatment at national or sub-national scale through micro-planning usually requires higher-resolution spatial data. A small-area mapping approach to snakebite has been pioneered in Costa Rica by Hansson et al. (2013) who modelled realistic travel time to health facilities and ambulance stations, together with habitat suitability maps for Bothrops asper, to identify populations with need of improved treatment access. A similar approach is currently being applied in Cameroon and Nepal to model vulnerable populations and optimize access to antivenom (Alcoba et al., 2021). In particularly difficult terrain such as the Amazons, understanding the extent to which the population is unable to rapidly access adequate care after a snakebite can trigger radically different solutions, such as antivenom delivery by drones (Meier and Bergelund, 2017).
As discussed earlier, models of spatial snakebite variation can be adequately tackled at 1 km or coarser resolution for some purposes, but accessibility modelling typically requires working at 100m or even 30m (e.g. Hierink et al., 2020) resolution. A finer raster resolution allows one to capture more realistically the landscape characteristics and infrastructures that can influence the movements of care-seeking patients. The recent availability of high-resolution datasets needed to model accessibility (openly accessible for most countries from sites such as Humanitarian Data Exchange, https://data.humdata.org/) has enabled a big push towards the application of geospatial accessibility models. However, a notable difficulty in many countries is to access a complete data set on the locations of health facilities. Recent projects have facilitated access and update of health facilities data (Maina et al., 2019; South et al., 2020), but knowing which facilities are effectively treating snakebite and have antivenom availability remains a challenge in most countries (see Potet et al., this issue). Notably, WHO is currently compiling data on health care facilities in several countries in Eastern and Western sub-Saharan Africa to provide a baseline for a targeted antivenom stockpiling project that, if successful could be expanded across this and other regions.
The nascent use of high-resolution accessibility modelling to better understand the population at risk of snakebite envenoming holds great promises. When data on spatial snakebite variation, spatial distributions of venomous snakes, and antivenom availability are more widely available, the modelling of vulnerable population coupled to accessibility modelling can be a game changer for planning and optimizing SBE-related care in affected countries. This also fits the scope of the “precision global health” (Flahault et al., 2020; Sheath et al., 2020) agenda that seeks, notably, to enhance effective resource allocation through use of high-resolution spatio-temporal data and innovative digital tools.
7. A changing world: the effect of land use change and climate change on human-snake interactions
Just as snakebite risk and incidence change with season, weather and time of day depending on human and snake activity patterns, they also show long-term temporal trends based on changes in climate, weather anomalies (Ediriweera et al., 2018) and human land use. Notably, this is not only of medical relevance, but also poses important challenges to conservation (Lara-Galván et al., 2020). As with other human-wildlife conflicts, the general public usually perceives snakes as a threat, but is less aware that they themselves also pose a threat to wildlife (Nyhus, 2016). Many snakes are International Union for Conservation of Nature (IUCN) listed (IUCN, 2020): out of those listed by WHO, three are considered critically endangered, nine endangered, 19 vulnerable, seven near threatened, 11 data deficient, and 85 have not been assessed. This does not yet include any species only listed under groupings such as ‘Micrurus species’, which have been suggested to be particularly vulnerable to climate change (Terribile et al., 2018) and achieving conservation goals can be difficult for organisms involved in human-wildlife conflicts (Madden, 2004).
Anthropogenic climate change will affect snake distributions and abundance, just as it affects many other organisms. Many animals, including snakes, are predicted to shift their ranges into higher latitudes as the climate warms, and correspondingly, contract their ranges at low latitudes (Behrooz et al., 2015; Hickling et al., 2006; Nori et al., 2014). For wide ranging species, which tend to have broader environmental tolerances (Pintor et al., 2015), this trend may not be of conservation concern, especially if the overall size of suitable area remains similar. However, from a human-snake interaction perspective, increases in snakebite risk can occur if snake ranges shift towards more densely populated areas or previously less exposed populations (Nori et al., 2014). Snake range shifts will also require changes in antivenom supply logistics as affected human populations shift with them. Such shifts will likely coincide with shifts in many vector-borne diseases (Campbell et al., 2015), resulting in a substantial challenge for global disease management (Lafferty, 2009). For range-restricted species and those associated with very specific habitats (e.g. mountain tops or specific vegetation; Behrooz et al., 2015; Freeman et al., 2018), climate change may pose a higher risk because limited dispersal ability and habitat fragmentations may hinder shifts in response to these changes (Terribile et al., 2018; Vasudev et al., 2015; Yousefi et al., 2015). Considering the concentration of range-restricted species within many taxa in the tropics (Pintor et al., 2015; Stevens, 1989), the threat of climate change to snake species and the threat of snakes to humans coincide in similar areas, i.e. in tropical developing countries. Understanding how snake ranges will change is crucial to future-proof snakebite management tactics.
Climate change may also affect snake abundance and activity patterns at a more local scale, thereby increasing snakebite risk. However, as we barely understand current patterns in snake abundance and activity, further research is urgently needed to assess how patterns will change in the future. For example, snake abundance and activity may increase locally because of longer warm or rainy seasons (DeGregorio et al., 2015; Ediriweera et al., 2018; Moreno-Rueda et al., 2009), breeding seasons could shift or reproductive output could change (Brown and Shine, 2007; Halupka and Halupka, 2017; Henle et al., 2008; Najmanová and Adamík, 2009), warmer temperatures could make snakes more active and likely to bite (Ediriweera et al., 2018; Schieffelin and de Queiroz, 1991) or snakes could change their daily activity patterns to make best use of favorable temperatures (Gordon et al., 2010; Levy et al., 2019). This could lead to increased human exposure, exposure at different times of day, or in different seasons. Climate change is also predicted to lead to more extreme weather anomalies (Mirza, 2003; Seneviratne et al., 2012; Stott, 2016). Weather anomalies (e.g. in maximum relative humidity) have been shown to coincide with changes in snakebite prevalence (Ediriweera et al., 2018).
Another dynamic aspect of human-snake interactions is human land use and how it changes in response to population growth, infrastructure development, changes in resource exploitation, or expansion of farming systems (Lamarque et al., 2009; Nyhus, 2016). For example, snakebite is usually rarer in densely populated urban areas (Ediriweera et al., 2016) and urbanization could lead to local decreases in snakebite prevalence. On the other hand, snakebite incidence in rural farming systems is high (Hansson et al., 2010, 2013; León-Núñez et al., 2020; Suraweera et al., 2020) and varies amongst different crops (Goldstein et al., 2021). Certain crop expansion and changes in farming practices could lead to increased snakebite prevalence, while mechanization of farming practices could, conversely, reduce exposure to snakes. Human-wildlife conflict also often increases with deforestation (Lamarque et al., 2009; Schneider et al., 2021) and in border-country to remnant forests and protected areas (Hansson et al., 2013; Sharma et al., 2020) because animals are forced to leave their natural habitat and use anthropogenic landscapes. For species that are incapable of using anthropogenic landscapes, this might lead to decreases in suitable habitat and short term increases in human encounters as they search for new suitable habitat (Acharya et al., 2017; Distefano, 2005). For species that profit from human landscapes or adapt easily to modified landscapes, it likely leads to increases in population numbers and long term increases in human exposure (Arias-Rodríguez and Gutiérrez, 2020; Löwenborg et al., 2010; Urbina-Cardona et al., 2008). Consequences for snakebite management are likely complex and depend on the species composition of any given landscape as well as the type and spatial patterns in land use change (e.g. broad scale conversion of natural habitats versus changes in patchiness in mosaic landscapes, proximity to protected areas, etc.; Acharya et al., 2017). Land use change itself is difficult to predict because it is based on complex drivers such as human decision-making processes and government policies (Hurtt et al., 2020; Li et al., 2017; Veldkamp and Lambin, 2001; Xie et al., 2014). However, there is a wide suite of literature and methods on land use change that could be integrated into efforts to manage snakebite into the future (Veldkamp and Lambin, 2001).
A complete review of the literature on land use change is out of the scope of this article, however, some examples are worth discussing. For example (Amici et al., 2017), used machine-learning algorithms to estimate the likelihood of land cover change based on previously observed conversion patterns. This approach is comparably low-effort because it uses existing satellite imagery, land use classifications and freely available spatial predictors in combination with well-established machine learning approaches. At the other end of the effort-scale are methods that document detailed decision-making patterns by individual landowners and governments to predict land conversion probabilities, often in combination with a land suitability analysis (Hurtt et al., 2020; Li et al., 2017; Veldkamp and Lambin, 2001; Xie et al., 2014). Similarly complex models have recently been developed at global scales, for example land use change for 2050 and 2100 for land types such as forests, grasslands, croplands, urban, and bare areas at 1 km resolution (Li et al., 2017), or historic and future land use classification from 850 to 2100 at 25 km (Hurtt et al., 2020). Existing predictions of land use change could be used to describe aspects relevant to human-snake interactions. For example, areas that have a high potential for smallholder-irrigated agriculture or are predicted to be changed to cropland have a higher chance of being converted to cropping systems that may increase human-snake interactions. However, for many regions, models of land use changes specifically relevant to human-snake interactions do not yet exist or not at suitable scales. Creation of new land use change probability maps that estimate change in specific parameters that might affect spatial snakebite variation would be highly beneficial. This is a major task but could feed into many other humanitarian aid efforts and even guide land use change planning and protected area management to help avoid worst-case scenarios for humans as well as for biodiversity. Ultimately, such land use change models, snake ENMs, snakebite incidence models, and analyses of healthcare accessibility could all feed into ‘multiple objective planning’ research, which aims to find best compromises for biodiversity, cultural, health, and economic objectives during land development planning (Álvarez-Romero et al., 2021).
8. Increasing the spatial resolution of venom variation to inform antivenom production and use
Snake venoms are sophisticated and complex mixtures of proteins that play important roles in prey acquisition and, to a lesser degree, self-defense (Daltry et al., 1996; Kazandjian et al., 2021). For many snakes, venoms are the primary mode of securing prey and hence have undergone strong selection pressures to function optimally depending on prey type and habitat (Healy et al., 2019; Sunagar and Moran, 2015), and to fulfill the specific function required (e.g. prey paralysis, digestion; (Fry, 2015; Fry et al., 2012; Kardong, 1982). Prey type, availability, and snake habitat varies geographically, especially for wide-ranging snakes (Daltry et al., 1996): consequently, different snake populations often evolve to have different arsenals of venom proteins between different geographic locations to optimize prey acquisition (Strickland et al., 2018). For example, pooled venom samples of Bitis arietans, a medically important, wide-ranging species in Africa with substantial phylogeographic differentiation (Barlow et al., 2013), vary in their protein profile, antibody cross-reactivity, and enzyme activity between Saudi Arabia, Nigeria, Ghana, Malawi, Tanzania, and Zimbabwe (Currier et al., 2010). Venoms of additional category 1 species Calloselasma rhodostoma, Bothrops asper, Bothrops atrox, and Crotalus scutulatus, vary substantially across their observed ranges (Alape-Girón et al., 2008; Daltry et al., 1996; Sousa et al., 2018; Strickland et al., 2018; Zancolli et al., 2019). Similarly, geographic variation in venom composition and immunology varies geographically in all ‘Big Four’ snake species of India (Echis carinatus, Naja naja, Daboia russelii, and Bungarus caeruleus which has the highest snakebite mortality in the World (Kalita et al., 2018; Kasturiratne et al., 2008; Mukherjee, 2020; Oh et al., 2017; Patra and Mukherjee, 2020; Pla et al., 2019; Senji Laxme et al., 2021a; Senji Laxme et al., 2021b, Fig. 5). Such geographic variation, along with ontogenetic, intra-population, and other forms of venom variation, has important consequences for snakebite management: antivenom efficacy can vary amongst localities depending on which populations were originally used for antivenom production and differences in enzyme activity can cause different clinical manifestations of envenomation (Casewell et al., 2014; Chippaux et al., 1991; Warrell, 1997).
Fig. 5.
Sample locations for studies on the geographic variation in venom composition in the ‘Big Four’ snakes across the Indian sub-continent: the Indian spectacled cobra (Naja naja; Mukherjee et al., 2020), the Indian krait (Bungarus caeruleus; Oh et al., 2017), the saw-scaled viper (Echis carinatus; Patra et al., 2020), and the Russell's viper (Daboia russelii; Pla et al., 2019). Background shows mean vegetation greenness (fraction photosynthetic active radiation; https://land.copernicus.eu/global/products/fapar) with greener areas shown as darker shades of grey.
Antivenoms are produced using pooled venoms from individuals of each species whose venoms they are designed to neutralize. It is, therefore, paramount that venoms used in antivenom production adequately represent the natural variation found across the geographic region where the antivenom will be used, to ensure their specificity and generality (Chippaux et al., 1991). In reality, however, this is rarely the case. Venom for antivenom production often comes from captive snake populations rather than being collected in the field (WHO, 2010a). In either case, the origin of these populations is usually of an opportunistic rather than planned nature. Furthermore, many snakes have no specific antivenom produced against them at all (Longbottom et al., 2018) and their bites are treated with antivenoms developed against related species, which is problematic since intra-genus venom variation can be substantial and is often as poorly understood as intraspecific variation (Queiroz et al., 2008). Part of the reason for these shortcomings is that it is difficult and expensive to obtain representative venom samples from all species and populations across large geographic regions. The other reason is that we simply do not have a good understanding of how venom composition varies geographically within most species (and sometimes amongst different species) and, therefore, cannot chose representative venom collection localities objectively. A resulting known unknown is that we can rarely say where an antivenom is effective or ineffective because of limited efficacy testing (Keyler et al., 2013; WHO, 2010a).
WHO's ‘guidelines for the production and validation of antivenoms’ outline solutions to the issue of poorly tested antivenoms with poorly documented production methodologies (WHO, 2010a, 2018): recommendations include the creation of region-specific polyvalent antivenoms, the careful consideration of appropriate venoms used in antivenom production, pre-clinical tests of neutralization efficiency of relevant region-specific venoms, and traceability of venom batches as well as consistency amongst batches. Despite ongoing efforts from WHO to test the quality of available antivenoms, poor quality antivenoms still dominate the current market. To put WHO's guidelines into broader practice, we require detailed studies of geographic venom variation, especially for snakes with large geographic ranges. As mentioned above, geographic venom variation has been studied in some snakes but mostly at a relatively coarse resolution. Venom is usually compared between different populations from extreme corners of a snake's distribution or from a subset of countries or states the species occupies (Chang et al., 2013; Currier et al., 2010; Mukherjee, 2020; Oh et al., 2017; Pla et al., 2019; Sousa et al., 2018). Few studies have comprehensively assessed venom variation at a fine-scale across the whole range of a species (Daltry et al., 1996; Strickland et al., 2018). Overall, our understanding of fine-scale geographic venom variation is limited. For example, the venom of a hypothetical species may be different between three populations P1, P2, and P3 (Fig. 6). These populations may represent three distinct clades that each cover a third of the species' distribution. Alternatively, venom composition may change gradually between the populations and be slightly different at any given location. Another possibility is that some population are restricted to small areas delineated by geographic boundaries to gene flow or that each have a distinct habitat type, while others are wide-ranging. There may even be additional distinct venom lineages (e.g. isolated island lineages) that have not yet been discovered and whose venom is not neutralized by antivenom based on the three known lineages. Lastly, different venom lineages can occur throughout a snake's distribution based on fine-scale environmental patterns (Strickland et al., 2018; Zancolli et al., 2019) or diversity of venom expression within a population (Pintor et al., 2011) instead of occupying distinct parts of the distribution.
Fig. 6.
Example of how different hypothetical intraspecific venom lineages or venom expression types could be distributed within a species' overall range. A: Location of the sampled lineages P1, P2, and P3; B: Each venom lineage may occur throughout the species' distribution (wide-spread diversity in expression of venom types); C: geographically distinct lineages could occupy similar proportions of the species' range; D: venom composition could change gradually between lineages; E: some lineages could be locally restricted because of boundaries to gene flow (thick black lines) or different sized areas of distinct habitat types relevant to venom expression; F: additional unsampled lineages may be present, such as isolated island [P4] or distinct habitat fragment [P5] lineages.
A good understanding of geographic venom variation can benefit snakebite management in a multitude of ways (Chippaux et al., 1991; Fry et al., 2003; Senji Laxme et al., 2021b). Firstly, it allows us to determine where current antivenoms are likely to work based on the origin of venom used for their production (Senji Laxme et al., 2021b). Potential gaps in efficacy can be identified and used to target additional venom collection for efficacy assessments or new antivenom development where necessary. Secondly, new antivenom regions could be defined based on the boundaries of known venom lineages and region-specific antivenoms produced to maximize efficacy and minimize required volumes (Keyler et al., 2013). Thirdly, studies on taxonomy and on drivers of venom evolution could profit from the observed patterns and use them to predict likely patterns in variation for snakes that have not yet been studied. This last point is particularly important considering the high cost, effort, and risk involved in surveying snake populations across vast, remote, and often politically unstable regions.
Distribution estimates based on predictive models could function as a basis for venom sampling efforts. For example, venom lineages may be similar across continuous patches of suitable habitat, while low suitability could function as a geographic barrier separating distinct lineages. Individual sampling locations from each suitable habitat patch (at appropriate scales) could be prioritized for venom collection, followed by more fine-scale collection efforts if resources allow (Fig. 7). Recent studies have used ENMs to estimate the distribution of individual genetic lineages within a clade based on cost-distance from known locations (Rosauer et al., 2015, 2016) and a similar approach could be used to estimate the distribution of venom lineages. In crisis scenarios where a new representative antivenom needs to be produced quickly or resources are limited, distinct suitable habitat patches could even be used as a proxy for potential venom lineages needed for representative venom collection and verified later (Fig. 7). Alternatively, distinct venom lineages can be modelled separately to study environmental drivers of venom variation (Strickland et al., 2018). Care must be taken to not confuse genetic lineages with venom lineages, as geographically distinct venom lineages have been shown to occur even within distinct genetic subpopulations (Strickland et al., 2018; Zancolli et al., 2019). Note that there are many more challenges involved in the improvement of antivenoms that are out of the scope of this article. Only the spatial components have been discussed here.
Fig. 7.
Example of how the species Bungarus fasciatus might be split up (thick black lines) into potential venom lineages based on perceived gaps in its distribution and known dispersal barriers (e.g. oceans) using (A) known occurrence records (red dots) and expert derived range estimates (pink shaded area) or, alternatively, using (B) habitat suitability estimates to detect potential distribution gaps.
9. Conclusion
Successful snakebite mitigation and management requires a fine-scale understanding of spatial patterns in snake distributions, snakebite incidence, human population vulnerability, and medical infrastructure globally. Considerable efforts must be taken to collect additional data within these categories and to streamline data integration and collaboration between governmental bodies, the scientific community and the general public. Only then can sophisticated spatio-temporal analysis methods be applied to accurately predict spatio-temporal variation, which will inform successful on-ground management and resource allocation. Until such systems are implemented, interim solutions can function as preliminary means to guide actions. Existing data collection and model methods in example countries can be expanded to surrounding regions. Research on snake biology and human-snake interactions can add value to existing models. Citizen science projects can test-run targeted elicitation of data collection in under-sampled areas using novel vetting protocols, possibly including image recognition. Lastly, snake conservation, education, and land use management can mitigate potential future increases in human-snake conflict.
We have outlined knowledge gaps and approaches to reduce them for a wide variety of spatial components of the global snakebite crisis. The key steps needed for progress are summarized from a practical, as well as academic perspective, in Text Box 1. However, successful snakebite management and prevention is influenced by many other, non-spatial factors that are discussed elsewhere in this special issue. These include topics such as antivenom production and quality control, community engagement strategies, mobilization of financial resources, improvements in snakebite first aid, medical personnel training, and medical protocols, amongst others.
Text Box 1. Recommended priorities for spatial snakebite research and management:
-
•
Streamline collection of data on snake occurrences & snakebite incidence
-
•
Set up pipelines for data vetting, data integration, and expert collaboration
-
•
Create globally consistent models of current and future snake distributions, snakebite risk, and snakebite incidence, under consideration of climate change, land use change, and seasonal variability
-
•
Study patterns in venomous snake abundance, activity, and population dynamics
-
•
Set up spatially optimized land use strategies and protected area networks that minimize human-snake conflict
-
•
Establish spatial database of health care infrastructure and vulnerable populations
-
•
Set up spatially optimized antivenom distribution networks
-
•
Determine fine-scale patterns in and evolutionary drivers of spatial venom variation
Alt-text: Text Box 1
We hope that this review will motivate future research on the topic, promoting additional transdisciplinary collaboration and innovation to expand the information and methods suggested here. The gap between traditional epidemiology, ecology, conservation biology, and information technology is worth narrowing to unite strengths against snakebite.
Credit author statement
Anna FV Pintor: Conceptualization, Writing – original draft, Writing – review & editing, Visualization, Methodology. Nicolas Ray: Writing – original draft, Writing – review & editing, Methodology. Joshua Longbottom: Writing – review & editing, Methodology. Carlos A Bravo-Vega: Writing – original draft, Writing – review & editing, Methodology. Masoud Yousef: Writing- Reviewing and Editing, Methodology. Kris A Murray: Writing- Reviewing and Editing, Methodology. Dileepa S Ediriweera: Writing- Reviewing and Editing, Visualization, Methodology. Peter J Diggle: Writing- Reviewing and Editing, Methodology.
We, the author, declare that this manuscript complies with all legal and ethical standards required by Toxicon: X. No human or animal experiments were conducted as part of this manuscript.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
We thank the Welcome Trust, London, UK (grant 222215/Z/20/Z to World Health. Organization) and the World Health Organization, Geneva, Switzerland for financial support. We also thank James Cook University, Cairns, Australia for technical support. KAM would like to acknowledge Joint Centre funding from the UK Medical Research Council (MRC) and Department for International Development (MR/R015600/1) and an MRC Global Challenges Research Fund award (MP/P024513/1).
Handling Editor: Raymond Norton
References
- Acharya K.P., Paudel P.K., Jnawali S.R., Neupane P.R., Koehl M. Can forest fragmentation and configuration work as indicators of human–wildlife conflict? Evidences from human death and injury by wildlife attacks in Nepal. Ecol. Indicat. 2017;80:74–83. [Google Scholar]
- Ahmed S., Adams A.M., Islam R., Hasan S.M., Panciera R. Impact of traffic variability on geographic accessibility to 24/7 emergency healthcare for the urban poor: a GIS study in Dhaka, Bangladesh. PloS One. 2019;14 doi: 10.1371/journal.pone.0222488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ALA . 2021. ALA Open Access to Australia's Biodiversity Data.https://www.ala.org.au/ 15/3/2021. [Google Scholar]
- Alape-Girón A., Sanz L., Escolano J., Flores-Diaz M., Madrigal M., Sasa M., Calvete J.J. Snake venomics of the lancehead pitviper Bothrops asper: geographic, individual, and ontogenetic variations. J. Proteome Res. 2008;7:3556–3571. doi: 10.1021/pr800332p. [DOI] [PubMed] [Google Scholar]
- Alcoba G., Ochoa C., Babo Martins S., Ruiz de Castañeda R., Bolon I., Wanda F., Comte E., Subedi M., Shah B., Ghimire A. Novel transdisciplinary methodology for cross-sectional analysis of snakebite epidemiology at national scale. PLoS Neglected Trop. Dis. 2021;15 doi: 10.1371/journal.pntd.0009023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Álvarez-Romero J.G., Kiatkoski Kim M., Pannell D., Douglas M.M., Wallace K., Hill R., Adams V.M., Spencer-Cotton A., Kennard M., Pressey R.L. Multi-objective planning in northern Australia: co-benefits and trade-offs between environmental, economic, and cultural outcomes. Final report to the Australian Department of Agriculture, Water and the Environment. 2021 [Google Scholar]
- Amici V., Marcantonio M., La Porta N., Rocchini D. A multi-temporal approach in MaxEnt modelling: a new frontier for land use/land cover change detection. Ecol. Inf. 2017;40:40–49. [Google Scholar]
- Anand S., Radhakrishna S. Investigating trends in human-wildlife conflict: is conflict escalation real or imagined? J. Asia Pac. Bus. 2017;10:154–161. [Google Scholar]
- Angarita-Gerlein D., Bravo-Vega C., Cruz C., Forero-Muñoz N., Navas-Zuloaga M., Umaña-Caro J. Snakebite dynamics in Colombia: effects of precipitation seasonality on incidence. IBIO4299 International Research Experience For Students IRES. 2017 [Google Scholar]
- Araújo M.B., New M. Ensemble forecasting of species distributions. Trends Ecol. Evol. 2007;22:42–47. doi: 10.1016/j.tree.2006.09.010. [DOI] [PubMed] [Google Scholar]
- Arctos . 2021. Arctos Collaborative Collection Management Solution.https://arctosdb.org/ [Google Scholar]
- Arias-Rodríguez J., Gutiérrez J.M. Circumstances and consequences of snakebite envenomings: a qualitative study in South-Eastern Costa Rica. Toxins. 2020;12:45. doi: 10.3390/toxins12010045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asadi A., Montgelard C., Nazarizadeh M., Moghaddasi A., Fatemizadeh F., Simonov E., Kami H.G., Kaboli M. Evolutionary history and postglacial colonization of an Asian pit viper (Gloydius halys caucasicus) into Transcaucasia revealed by phylogenetic and phylogeographic analyses. Sci. Rep. 2019;9:1–16. doi: 10.1038/s41598-018-37558-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banes Kieran, Ngari Cecelia, Parkurito Stanley, Wood Leo, Otundo Denis, Harrison Robert, George Oluoch, O, Baker Clare. Delays, fears and training needs: Perspectives of health workers on clinical management of snakebite revealed by a qualitative study in Kitui County, Kenya. Toxicon: X. 2021 doi: 10.1016/j.toxcx.2021.100078. In this issue. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barasa E.W., Ouma P.O., Okiro E.A. Assessing the hospital surge capacity of the Kenyan health system in the face of the COVID-19 pandemic. PloS One. 2020;15 doi: 10.1371/journal.pone.0236308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barlow A., Baker K., Hendry C.R., Peppin L., Phelps T., Tolley K.A., Wüster C.E., Wüster W. Phylogeography of the widespread African puff adder (B itis arietans) reveals multiple Pleistocene refugia in southern Africa. Mol. Ecol. 2013;22:1134–1157. doi: 10.1111/mec.12157. [DOI] [PubMed] [Google Scholar]
- Barve V. Discovering and developing primary biodiversity data from social networking sites: a novel approach. Ecol. Inf. 2014;24:194–199. [Google Scholar]
- Beattie K., Olson E., Kissui B., Kirschbaum A., Kiffner C. Predicting livestock depredation risk by African lions (Panthera leo) in a multi-use area of northern Tanzania. Eur. J. Wildl. Res. 2020;66:1–14. [Google Scholar]
- Behrooz R., Kaboli M., Nourani E., Ahmadi M., Shabani A.A., Yousefi M., Asadi A., Rajabizadeh M. Habitat modeling and conservation of the endemic latifi's viper (montivipera latifii) in lar national park, northern Iran. Herpetol. Conserv. Biol. 2015;10:572–582. [Google Scholar]
- BISON . 2021. Biodiversity Information Serving Our Nation (BISON) - Explore & Download North American Species Occurrence Data & Maps.https://bison.usgs.gov/#home 15/3/2021. [Google Scholar]
- Blouin‐Demers G., Weatherhead P.J. An experimental test of the link between foraging, habitat selection and thermoregulation in black rat snakes Elaphe obsoleta obsoleta. J. Anim. Ecol. 2001;70:1006–1013. [Google Scholar]
- Boakes E.H., McGowan P.J., Fuller R.A., Chang-qing D., Clark N.E., O'Connor K., Mace G.M. Distorted views of biodiversity: spatial and temporal bias in species occurrence data. PLoS Biol. 2010;8 doi: 10.1371/journal.pbio.1000385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bravo-Vega C.A., Cordovez J.M., Renjifo-Ibáñez C., Santos-Vega M., Sasa M. Estimating snakebite incidence from mathematical models: a test in Costa Rica. PLoS Neglected Trop. Dis. 2019;13 doi: 10.1371/journal.pntd.0007914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braz A.G., de Viveiros Grelle C.E., de Souza Lima Figueiredo M., Weber M.d.M. Interspecific competition constrains local abundance in highly suitable areas. Ecography. 2020;43:1560–1570. [Google Scholar]
- Breiman L. Random forests. Mach. Learn. 2001;45:5–32. [Google Scholar]
- Brito J.C., Santos X., Pleguezuelos J.M., Sillero N. Inferring evolutionary scenarios with geostatistics and geographical information systems for the viperid snakes Vipera latastei and Vipera monticola. Biol. J. Linn. Soc. 2008;95:790–806. [Google Scholar]
- Brown G.P., Shine R. Rain, prey and predators: climatically driven shifts in frog abundance modify reproductive allometry in a tropical snake. Oecologia. 2007;154:361–368. doi: 10.1007/s00442-007-0842-8. [DOI] [PubMed] [Google Scholar]
- Brown J.H. On the relationship between abundance and distribution of species. Am. Nat. 1984;124:255–279. [Google Scholar]
- Burbrink F.T., Guiher T.J. Considering gene flow when using coalescent methods to delimit lineages of North American pitvipers of the genus Agkistrodon. Zool. J. Linn. Soc. 2015;173:505–526. [Google Scholar]
- Campbell L.P., Luther C., Moo-Llanes D., Ramsey J.M., Danis-Lozano R., Peterson A.T. Climate change influences on global distributions of dengue and chikungunya virus vectors. Phil. Trans. Biol. Sci. 2015;370:20140135. doi: 10.1098/rstb.2014.0135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carter N., Williamson M.A., Gilbert S., Lischka S.A., Prugh L.R., Lawler J.J., Metcalf A.L., Jacob A.L., Castro A.J., Sage A. Integrated spatial analysis for human-wildlife coexistence in the American West. Environ. Res. Lett. 2020;15(2) doi: 10.1088/1748-9326/ab60e1. [DOI] [Google Scholar]
- Casewell N.R., Jackson T.N., Laustsen A.H., Sunagar K. Causes and consequences of snake venom variation. Trends Pharmacol. Sci. 2020;41(8):570–581. doi: 10.1016/j.tips.2020.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casewell N.R., Wagstaff S.C., Wüster W., Cook D.A., Bolton F.M., King S.I., Pla D., Sanz L., Calvete J.J., Harrison R.A. Medically important differences in snake venom composition are dictated by distinct postgenomic mechanisms. Proc. Natl. Acad. Sci. Unit. States Am. 2014;111:9205–9210. doi: 10.1073/pnas.1405484111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandler M., See L., Copas K., Bonde A.M., López B.C., Danielsen F., Legind J.K., Masinde S., Miller-Rushing A.J., Newman G. Contribution of citizen science towards international biodiversity monitoring. Biol. Conserv. 2017;213:280–294. [Google Scholar]
- Chang H.-C., Tsai T.-S., Tsai I.-H. Functional proteomic approach to discover geographic variations of king cobra venoms from Southeast Asia and China. J. Proteomics. 2013;89:141–153. doi: 10.1016/j.jprot.2013.06.012. [DOI] [PubMed] [Google Scholar]
- Chaves L.F., Chuang T.-W., Sasa M., Gutiérrez J.M. Snakebites are associated with poverty, weather fluctuations, and El Niño. Sci. Adv. 2015;1 doi: 10.1126/sciadv.1500249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y.N., Schmitz M.M., Serbanescu F., Dynes M.M., Maro G., Kramer M.R. Geographic access modeling of emergency obstetric and neonatal care in Kigoma Region, Tanzania: transportation schemes and programmatic implications. Glob. Health: Sci. Prac. 2017;5:430–445. doi: 10.9745/GHSP-D-17-00110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chippaux J.-P. Estimate of the burden of snakebites in sub-Saharan Africa: a meta-analytic approach. Toxicon. 2011;57:586–599. doi: 10.1016/j.toxicon.2010.12.022. [DOI] [PubMed] [Google Scholar]
- Chippaux J.-P. Epidemiology of snakebites in Europe: a systematic review of the literature. Toxicon. 2012;59:86–99. doi: 10.1016/j.toxicon.2011.10.008. [DOI] [PubMed] [Google Scholar]
- Chippaux J.-P. Incidence and mortality due to snakebite in the Americas. PLoS Neglected Trop. Dis. 2017;11 doi: 10.1371/journal.pntd.0005662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chippaux J.-P., Williams V., White J. Snake venom variability: methods of study, results and interpretation. Toxicon. 1991;29:1279–1303. doi: 10.1016/0041-0101(91)90116-9. [DOI] [PubMed] [Google Scholar]
- Chippaux J.P. Snake-bites: appraisal of the global situation. Bull. World Health Organ. 1998;76:515. [PMC free article] [PubMed] [Google Scholar]
- Colasanti R. Discussions of the possible use of neural network algorithms in ecological modeling. Bin. Comput. Microbiol. 1991;3:13–15. [Google Scholar]
- Currier R.B., Harrison R.A., Rowley P.D., Laing G.D., Wagstaff S.C. Intra-specific variation in venom of the African Puff Adder (Bitis arietans): differential expression and activity of snake venom metalloproteinases (SVMPs) Toxicon. 2010;55:864–873. doi: 10.1016/j.toxicon.2009.12.009. [DOI] [PubMed] [Google Scholar]
- da Silva Souza A., Sachett J.d.A.G., Alcântara J.A., Freire M., Alecrim M.d.G.C., Lacerda M., de Lima Ferreira L.C., Fan H.W., de Souza Sampaio V., Monteiro W.M. Snakebites as cause of deaths in the western Brazilian Amazon: why and who dies? Deaths from snakebites in the Amazon. Toxicon. 2018;145:15–24. doi: 10.1016/j.toxicon.2018.02.041. [DOI] [PubMed] [Google Scholar]
- Dallas T., Decker R.R., Hastings A. Species are not most abundant in the centre of their geographic range or climatic niche. Ecol. Lett. 2017;20:1526–1533. doi: 10.1111/ele.12860. [DOI] [PubMed] [Google Scholar]
- Dallas T.A., Hastings A. Habitat suitability estimated by niche models is largely unrelated to species abundance. Global Ecol. Biogeogr. 2018;27:1448–1456. [Google Scholar]
- Daltry J.C., Wüster W., Thorpe R.S. Diet and snake venom evolution. Nature. 1996;379:537–540. doi: 10.1038/379537a0. [DOI] [PubMed] [Google Scholar]
- DeGregorio B.A., Westervelt J.D., Weatherhead P.J., Sperry J.H. Indirect effect of climate change: shifts in ratsnake behavior alter intensity and timing of avian nest predation. Ecol. Model. 2015;312:239–246. [Google Scholar]
- Dehghani R., Fathi B., Shahi M.P., Jazayeri M. Ten years of snakebites in Iran. Toxicon. 2014;90:291–298. doi: 10.1016/j.toxicon.2014.08.063. [DOI] [PubMed] [Google Scholar]
- Delamater P.L., Messina J.P., Shortridge A.M., Grady S.C. Measuring geographic access to health care: raster and network-based methods. Int. J. Health Geogr. 2012;11:1–18. doi: 10.1186/1476-072X-11-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desmond-Hellmann S. Progress lies in precision. Science. 2016;353(6301):731. doi: 10.1126/science.aai7598. [DOI] [PubMed] [Google Scholar]
- Di Cola V., Chiaraviglio M. Establishing species' environmental requirements to understand how the southernmost species of South American pitvipers (Bothrops, Viperidae) are distributed: a niche‐based modelling approach. Austral Ecol. 2011;36:90–98. [Google Scholar]
- Diniz‐Filho J.A.F., Mauricio Bini L., Fernando Rangel T., Loyola R.D., Hof C., Nogués‐Bravo D., Araújo M.B. Partitioning and mapping uncertainties in ensembles of forecasts of species turnover under climate change. Ecography. 2009;32:897–906. [Google Scholar]
- Distefano E. Food and Agricultural Organization of the United Nations (FAO), Sustainable Agriculture and Rural Development Initiative (SARDI), Rome, Italy. Available from: FAO Corporate Document Repository. 2005. Human-Wildlife Conflict worldwide: collection of case studies, analysis of management strategies and good practices.http://www.fao.org/documents [Google Scholar]
- Durso A., Bolon I., Kleinhesselink A., Mondardini M., Fernandez-Marquez J., Gutsche-Jones F., Gwilliams C., Tanner M., Smith C., Wüster W. Vol. 8. Royal Society Open Science; 2021. Crowdsourcing snake identification with online communities of professional herpetologists and avocational snake enthusiasts; p. 201273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ealy M.J., Fleet R.R., Rudolph D.C. Diel activity patterns of the Louisiana pine snakes (Pituophis ruthveni) in eastern Texas. Texs J. Sci. 2004;56(4):383–394. [Google Scholar]
- Ebener S., Stenberg K., Brun M., Monet J.-P., Ray N., Sobel H.L., Roos N., Gault P., Conlon C.M., Bailey P. Proposing standardised geographical indicators of physical access to emergency obstetric and newborn care in low-income and middle-income countries. BMJ global health. 2019;4 doi: 10.1136/bmjgh-2018-000778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ediriweera D.S., Diggle P.J., Kasturiratne A., Pathmeswaran A., Gunawardena N.K., Jayamanne S.F., Isbister G.K., Dawson A., Lalloo D.G., de Silva H.J. Evaluating temporal patterns of snakebite in Sri Lanka: the potential for higher snakebite burdens with climate change. Int. J. Epidemiol. 2018;47:2049–2058. doi: 10.1093/ije/dyy188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ediriweera D.S., Kasthuriratne A., Pathmeswaran A., Gunawardene N.K., Jayamanne S.F., Murray K., Iwamura T., Lalloo D.G., de Silva H.J., Diggle P.J. Adjusting for spatial variation when assessing individual-level risk: a case-study in the epidemiology of snake-bite in Sri Lanka. PloS One. 2019;14 doi: 10.1371/journal.pone.0223021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ediriweera D.S., Kasturiratne A., Pathmeswaran A., Gunawardena N.K., Jayamanne S.F., Lalloo D.G., de Silva H.J. Health seeking behavior following snakebites in Sri Lanka: results of an island wide community based survey. PLoS Neglected Trop. Dis. 2017;11 doi: 10.1371/journal.pntd.0006073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ediriweera D.S., Kasturiratne A., Pathmeswaran A., Gunawardena N.K., Wijayawickrama B.A., Jayamanne S.F., Isbister G.K., Dawson A., Giorgi E., Diggle P.J. Mapping the risk of snakebite in Sri Lanka-a national survey with geospatial analysis. PLoS Neglected Trop. Dis. 2016;10 doi: 10.1371/journal.pntd.0004813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehrlén J., Morris W.F. Predicting changes in the distribution and abundance of species under environmental change. Ecol. Lett. 2015;18:303–314. doi: 10.1111/ele.12410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elith J., Graham H., P C., Anderson R., Dudík M., Ferrier S., Guisan A., Hijmans J., Huettmann R., Leathwick F.R., J, Lehmann A. Novel methods improve prediction of species' distributions from occurrence data. Ecography. 2006;29:129–151. [Google Scholar]
- Elith J., Leathwick J. 2017. Boosted Regression Trees for Ecological Modeling. R Documentation.https://cran.r-project.org/web/packages/dismo/vignettes/brt.pdf [Google Scholar]
- Elith J., Leathwick J.R., Hastie T. A working guide to boosted regression trees. J. Anim. Ecol. 2008;77:802–813. doi: 10.1111/j.1365-2656.2008.01390.x. [DOI] [PubMed] [Google Scholar]
- ESA Land cover CCI product user guide version 2. Tech. 2017 http://maps.elie.ucl.ac.be/CCI/viewer/download/ESACCI-LC-Ph2-PUGv2_2.0.pdf https://cds.climate.copernicus.eu/cdsapp#!/dataset/satellite-land-cover?tab=overview 15/3/2021. [Google Scholar]
- Escobar L.E., Craft M.E. Advances and limitations of disease biogeography using ecological niche modeling. Front. Microbiol. 2016;7:1174. doi: 10.3389/fmicb.2016.01174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Escobar L.E., Peterson A.T., Favi M., Yung V., Pons D.J., Medina-Vogel G. Ecology and geography of transmission of two bat-borne rabies lineages in Chile. PLoS Neglected Trop. Dis. 2013;7:e2577. doi: 10.1371/journal.pntd.0002577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans J.S., Murphy M.A., Holden Z.A., Cushman S.A. Springer; 2011. Modeling Species Distribution and Change Using Random Forest, Predictive Species and Habitat Modeling in Landscape Ecology; pp. 139–159. [Google Scholar]
- Ferro C., López M., Fuya P., Lugo L., Cordovez J.M., González C. Spatial distribution of sand fly vectors and eco-epidemiology of cutaneous leishmaniasis transmission in Colombia. PloS One. 2015;10 doi: 10.1371/journal.pone.0139391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ficetola G.F., Bonardi A., Sindaco R., Padoa‐Schioppa E. Estimating patterns of reptile biodiversity in remote regions. J. Biogeogr. 2013;40:1202–1211. [Google Scholar]
- Fick S.E., Hijmans R.J. WorldClim 2: new 1‐km spatial resolution climate surfaces for global land areas. Int. J. Climatol. 2017;37:4302–4315. [Google Scholar]
- Flahault A., Utzinger J., Eckerle I., Sheath D.J., de Castañeda R.R., Bolon I., Bempong N.-E., Andayi F. Precision global health for real-time action. Lancet Digital Health. 2020;2:e58–e59. doi: 10.1016/S2589-7500(19)30240-7. [DOI] [PubMed] [Google Scholar]
- Fox S., Rathuwithana A., Kasturiratne A., Lalloo D., De Silva H. Underestimation of snakebite mortality by hospital statistics in the Monaragala District of Sri Lanka. Trans. R. Soc. Trop. Med. Hyg. 2006;100:693–695. doi: 10.1016/j.trstmh.2005.09.003. [DOI] [PubMed] [Google Scholar]
- Freeman B.G., Scholer M.N., Ruiz-Gutierrez V., Fitzpatrick J.W. Climate change causes upslope shifts and mountaintop extirpations in a tropical bird community. Proc. Natl. Acad. Sci. Unit. States Am. 2018;115:11982–11987. doi: 10.1073/pnas.1804224115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fry B. Oxford University Press; 2015. Venomous Reptiles and Their Toxins: Evolution, Pathophysiology and Biodiscovery. [Google Scholar]
- Fry B.G., Casewell N.R., Wüster W., Vidal N., Young B., Jackson T.N. The structural and functional diversification of the Toxicofera reptile venom system. Toxicon. 2012;60:434–448. doi: 10.1016/j.toxicon.2012.02.013. [DOI] [PubMed] [Google Scholar]
- Fry B.G., Winkel K.D., Wickramaratna J.C., Hodgson W.C., Wüster W. Effectiveness of snake antivenom: species and regional venom variation and its clinical impact. J. Toxicol. Toxin Rev. 2003;22:23–34. [Google Scholar]
- Funk C., Peterson P., Landsfeld M., Pedreros D., Verdin J., Shukla S., Husak G., Rowland J., Harrison L., Hoell A. The climate hazards infrared precipitation with stations—a new environmental record for monitoring extremes. Sci. Data. 2015;2:1–21. doi: 10.1038/sdata.2015.66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuster B., Sánchez-Zapero J., Camacho F., García-Santos V., Verger A., Lacaze R., Weiss M., Baret F., Smets B. Quality assessment of PROBA-V LAI, fAPAR and fCOVER collection 300 m products of copernicus global land service. Rem. Sens. 2020;12:1017. [Google Scholar]
- GBIF . 2021. GBIF Global Biodiversity Information Facility - Free and Open Access to Biodiversity Data.https://www.gbif.org [Google Scholar]
- Genevieve L.D., Ray N., Chappuis F., Alcoba G., Mondardini M.R., Bolon I., de Castaneda R.R. vol. 12. 2018. (Participatory Approaches and Open Data on Venomous Snakes: A Neglected Opportunity in the Global Snakebite Crisis? PLoS Neglected Tropical Diseases). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldstein E., Erinjery J.J., Martin G., Kasturiratne A., Ediriweera D.S., de Silva H.J., Diggle P., Lalloo D.G., Murray K.A., Iwamura T. Integrating human behavior and snake ecology with agent-based models to predict snakebite in high risk landscapes. PLoS Neglected Trop. Dis. 2021;15 doi: 10.1371/journal.pntd.0009047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon C.E., Dickman C.R., Thompson M.B. What factors allow opportunistic nocturnal activity in a primarily diurnal desert lizard (Ctenotus pantherinus)? Comp. Biochem. Physiol. Mol. Integr. Physiol. 2010;156:255–261. doi: 10.1016/j.cbpa.2010.02.007. [DOI] [PubMed] [Google Scholar]
- Goswami V.R., Medhi K., Nichols J.D., Oli M.K. Mechanistic understanding of human–wildlife conflict through a novel application of dynamic occupancy models. Conserv. Biol. 2015;29:1100–1110. doi: 10.1111/cobi.12475. [DOI] [PubMed] [Google Scholar]
- Grassly N.C., Fraser C. Mathematical models of infectious disease transmission. Nat. Rev. Microbiol. 2008;6:477–487. doi: 10.1038/nrmicro1845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grego J.M. Generalized additive models. Encyclopedia of Environmetrics. 2006;2 [Google Scholar]
- Guisan A., Edwards T.C., Jr., Hastie T. Generalized linear and generalized additive models in studies of species distributions: setting the scene. Ecol. Model. 2002;157:89–100. [Google Scholar]
- Guisan A., Thuiller W. Predicting species distribution: offering more than simple habitat models. Ecol. Lett. 2005;8:993–1009. doi: 10.1111/j.1461-0248.2005.00792.x. [DOI] [PubMed] [Google Scholar]
- Guisan A., Tingley R., Baumgartner J.B., Naujokaitis‐Lewis I., Sutcliffe P.R., Tulloch A.I., Regan T.J., Brotons L., McDonald‐Madden E., Mantyka‐Pringle C. Predicting species distributions for conservation decisions. Ecol. Lett. 2013;16:1424–1435. doi: 10.1111/ele.12189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gül S. Potential distribution modeling and morphology of pelias barani (Böhme and joger, 1983) in Turkey. Asian Herpetological Res. 2015;6(3):206–212. doi: 10.16373/j.cnki.ahr.140079. [DOI] [Google Scholar]
- Gutiérrez J.M., Calvete J.J., Habib A.G., Harrison R.A., Williams D.J., Warrell D.A. Snakebite envenoming. Nature Rev. Disease Primers. 2017;3:1–21. doi: 10.1038/nrdp.2017.79. [DOI] [PubMed] [Google Scholar]
- Gutiérrez J.M., León G., Burnouf T. Antivenoms for the treatment of snakebite envenomings: the road ahead. Biologicals. 2011;39:129–142. doi: 10.1016/j.biologicals.2011.02.005. [DOI] [PubMed] [Google Scholar]
- Gutiérrez J.M., Warrell D.A., Williams D.J., Jensen S., Brown N., Calvete J.J., Harrison R.A., Initiative G.S. The need for full integration of snakebite envenoming within a global strategy to combat the neglected tropical diseases: the way forward. PLoS Neglected Trop. Dis. 2013;7 doi: 10.1371/journal.pntd.0002162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutiérrez J.M., Williams D., Fan H.W., Warrell D.A. Snakebite envenoming from a global perspective: towards an integrated approach. Toxicon. 2010;56:1223–1235. doi: 10.1016/j.toxicon.2009.11.020. [DOI] [PubMed] [Google Scholar]
- Habib A., Abubakar S. Factors affecting snakebite mortality in north-eastern Nigeria. Int. Health. 2011;3:50–55. doi: 10.1016/j.inhe.2010.08.001. [DOI] [PubMed] [Google Scholar]
- Habib A.G., Brown N.I. The snakebite problem and antivenom crisis from a health-economic perspective. Toxicon. 2018;150:115–123. doi: 10.1016/j.toxicon.2018.05.009. [DOI] [PubMed] [Google Scholar]
- Halilu S., Iliyasu G., Hamza M., Chippaux J.-P., Kuznik A., Habib A.G. Snakebite burden in Sub-Saharan Africa: estimates from 41 countries. Toxicon. 2019;159:1–4. doi: 10.1016/j.toxicon.2018.12.002. [DOI] [PubMed] [Google Scholar]
- Halupka L., Halupka K. The effect of climate change on the duration of avian breeding seasons: a meta-analysis. Proc. Biol. Sci. 2017;284:20171710. doi: 10.1098/rspb.2017.1710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamm N.A., Soares Magalhães R.J., Clements A.C. Earth observation, spatial data quality, and neglected tropical diseases. PLoS Neglected Trop. Dis. 2015;9 doi: 10.1371/journal.pntd.0004164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansdak S.G., Lallar K.S., Pokharel P., Shyangwa P., Karki P., Koirala S. A clinico-epidemiological study of snake bite in Nepal. Trop. Doct. 1998;28:223–226. doi: 10.1177/004947559802800412. [DOI] [PubMed] [Google Scholar]
- Hansson E., Cuadra S., Oudin A., de Jong K., Stroh E., Torén K., Albin M. Mapping snakebite epidemiology in Nicaragua–pitfalls and possible solutions. PLoS Neglected Trop. Dis. 2010;4:e896. doi: 10.1371/journal.pntd.0000896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansson E., Sasa M., Mattisson K., Robles A., Gutiérrez J.M. Using geographical information systems to identify populations in need of improved accessibility to antivenom treatment for snakebite envenoming in Costa Rica. PLoS Neglected Trop. Dis. 2013;7 doi: 10.1371/journal.pntd.0002009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison R.A., Gutiérrez J.M. Priority actions and progress to substantially and sustainably reduce the mortality, morbidity and socioeconomic burden of tropical snakebite. Toxins. 2016;8:351. doi: 10.3390/toxins8120351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison R.A., Hargreaves A., Wagstaff S.C., Faragher B., Lalloo D.G. Snake envenoming: a disease of poverty. PLoS Neglected Trop. Dis. 2009;3 doi: 10.1371/journal.pntd.0000569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hastie T.J., Tibshirani R. Generalized additive models: some applications. J. Am. Stat. Assoc. 1987;82:371–386. [Google Scholar]
- Hastie T.J., Tibshirani R.J. CRC press; 1990. Generalized Additive Models. [DOI] [PubMed] [Google Scholar]
- Healy K., Carbone C., Jackson A.L. Snake venom potency and yield are associated with prey‐evolution, predator metabolism and habitat structure. Ecol. Lett. 2019;22:527–537. doi: 10.1111/ele.13216. [DOI] [PubMed] [Google Scholar]
- Henle K., Dick D., Harpke A., Kühn I., Schweiger O., Settele J. Convention Council of Europe Publishing; Strasbourg, France: 2008. Climate Change Impacts on European Amphibians and Reptiles, Biodiversity and Climate Change: Reports and Guidance Developed under the Bern; pp. 225–305. [Google Scholar]
- HerpMapper HerpMapper. 2021. https://www.herpmapper.org/ 15/3/2021.
- Hickling R., Roy D.B., Hill J.K., Fox R., Thomas C.D. The distributions of a wide range of taxonomic groups are expanding polewards. Global Change Biol. 2006;12:450–455. [Google Scholar]
- Hierink F., Rodrigues N., Muñiz M., Panciera R., Ray N. Modelling geographical accessibility to support disaster response and rehabilitation of a healthcare system: an impact analysis of Cyclones Idai and Kenneth in Mozambique. BMJ open. 2020;10 doi: 10.1136/bmjopen-2020-039138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill C.M. Perspectives of “conflict” at the wildlife–agriculture boundary: 10 years on. Hum. Dimens. Wildl. 2015;20:296–301. [Google Scholar]
- Hurtt G.C., Chini L., Sahajpal R., Frolking S., Bodirsky B.L., Calvin K., Doelman J.C., Fisk J., Fujimori S., Klein Goldewijk K. Harmonization of global land use change and management for the period 850–2100 (LUH2) for CMIP6. Geosci. Model Dev. 2020;13:5425–5464. [Google Scholar]
- iDigBio . 2021. iDigBio Integrated Digitized Biocollections.https://www.idigbio.org/ [Google Scholar]
- Iliyasu G., Tiamiyu A.B., Daiyab F.M., Tambuwal S.H., Habib Z.G., Habib A.G. 2015. Effect of Distance and Delay in Access to Care on Outcome of Snakebite in Rural North-Eastern Nigeria. [PubMed] [Google Scholar]
- iNaturalist iNaturalist. 2021. https://www.inaturalist.org/) accesed 15/3/2021
- IPCC . Summary for policymakers. In: Shukla P.R., Skea J., Calvo Buendia E., Masson-Delmotte V., Pörtner H.-O., Roberts D.C., Zhai P., Slade R., Connors S., van Diemen R., Ferrat M., Haughey E., Luz S., Neogi S., Pathak M., Petzold J., Portugal Pereira J., Vyas P., Huntley E., Kissick K., Belkacemi M., Malley J., editors. Climate Change and Land: an IPCC Special Report on Climate Change, Desertification, Land Degradation, Sustainable Land Management, Food Security, and Greenhouse Gas Fluxes in Terrestrial Ecosystems. 2019. (in press) [Google Scholar]
- IUCN . 2020. The IUCN Red List of Threatened Species.https://www.iucnredlist.org Version 2020-3. [Google Scholar]
- Jacob S.T., Crozier I., Fischer W.A., Hewlett A., Kraft C.S., de La Vega M.-A., Soka M.J., Wahl V., Griffiths A., Bollinger L. Ebola virus disease. Nature Rev. Disease Primers. 2020;6:1–31. doi: 10.1038/s41572-020-0147-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jhala Y., Gopal R., Mathur V., Ghosh P., Negi H.S., Narain S., Yadav S.P., Malik A., Garawad R., Qureshi Q. Recovery of tigers in India: critical introspection and potential lessons. People Nat. 2021 [Google Scholar]
- Jiménez‐Valverde A., Aragón P., Lobo J.M. Deconstructing the abundance–suitability relationship in species distribution modelling. Global Ecol. Biogeogr. 2021;30:327–338. [Google Scholar]
- Jones B.A., Grace D., Kock R., Alonso S., Rushton J., Said M.Y., McKeever D., Mutua F., Young J., McDermott J. Zoonosis emergence linked to agricultural intensification and environmental change. Proc. Natl. Acad. Sci. Unit. States Am. 2013;110:8399–8404. doi: 10.1073/pnas.1208059110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joseph N.K., Macharia P.M., Ouma P.O., Mumo J., Jalang’o R., Wagacha P.W., Achieng V.O., Ndung’u E., Okoth P., Muñiz M. Spatial access inequities and childhood immunisation uptake in Kenya. BMC Publ. Health. 2020;20:1–12. doi: 10.1186/s12889-020-09486-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalita B., Mackessy S.P., Mukherjee A.K. Proteomic analysis reveals geographic variation in venom composition of Russell's Viper in the Indian subcontinent: implications for clinical manifestations post-envenomation and antivenom treatment. Expet Rev. Proteonomics. 2018;15:837–849. doi: 10.1080/14789450.2018.1528150. [DOI] [PubMed] [Google Scholar]
- Karabuva S., Vrkić I., Brizić I., Ivić I., Lukšić B. Venomous snakebites in children in southern Croatia. Toxicon. 2016;112:8–15. doi: 10.1016/j.toxicon.2016.01.057. [DOI] [PubMed] [Google Scholar]
- Kardong K.V. The evolution of the venom apparatus in snakes from colubrids to viperids and elapids. Mem. Inst. Butantan (Sao Paulo) 1982;46:105–118. [Google Scholar]
- Kasturiratne A., Pathmeswaran A., Wickremasinghe A.R., Jayamanne S.F., Dawson A., Isbister G.K., de Silva H.J., Lalloo D.G. The socio-economic burden of snakebite in Sri Lanka. PLoS Neglected Trop. Dis. 2017;11 doi: 10.1371/journal.pntd.0005647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasturiratne A., Wickremasinghe A.R., de Silva N., Gunawardena N.K., Pathmeswaran A., Premaratna R., Savioli L., Lalloo D.G., de Silva H.J. The global burden of snakebite: a literature analysis and modelling based on regional estimates of envenoming and deaths. PLoS Med. 2008;5:e218. doi: 10.1371/journal.pmed.0050218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kazandjian T.D., Petras D., Robinson S.D., van Thiel J., Greene H.W., Arbuckle K., Barlow A., Carter D.A., Wouters R.M., Whiteley G. Convergent evolution of pain-inducing defensive venom components in spitting cobras. Science. 2021;371:386–390. doi: 10.1126/science.abb9303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keyler D., Gawarammana I., Gutiérrez J.M., Sellahewa K., McWhorter K., Malleappah R. Antivenom for snakebite envenoming in Sri Lanka: the need for geographically specific antivenom and improved efficacy. Toxicon. 2013;69:90–97. doi: 10.1016/j.toxicon.2013.01.022. [DOI] [PubMed] [Google Scholar]
- Kiffner C., Schaal I., Cass L., Peirce K., Sussman O., Grueser A., Wachtel E., Adams H., Clark K., König H.J. Perceptions and realities of elephant crop raiding and mitigation methods. Conservation Sci. Practice. 2021:e372. [Google Scholar]
- Kim C., Tappis H., McDaniel P., Soroush M.S., Fried B., Weinberger M., Trogdon J.G., Lich K.H., Delamater P.L. National and subnational estimates of coverage and travel time to emergency obstetric care in Afghanistan: modeling of spatial accessibility. Health Place. 2020;66:102452. doi: 10.1016/j.healthplace.2020.102452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kretser H.E., Sullivan P.J., Knuth B.A. Housing density as an indicator of spatial patterns of reported human–wildlife interactions in Northern New York. Landsc. Urban Plann. 2008;84:282–292. [Google Scholar]
- Lafferty K.D. The ecology of climate change and infectious diseases. Ecology. 2009;90:888–900. doi: 10.1890/08-0079.1. [DOI] [PubMed] [Google Scholar]
- Laliberte A.S., Ripple W.J. Wildlife encounters by Lewis and Clark: a spatial analysis of interactions between Native Americans and wildlife. Bioscience. 2003;53:994–1003. [Google Scholar]
- Lamarque F., Anderson J., Fergusson R., Lagrange M., Osei-Owusu Y., Bakker L. Food and Agriculture Organization of the United Nations (FAO); 2009. Human-wildlife Conflict in Africa: Causes, Consequences and Management Strategies. [Google Scholar]
- Lara-Galván J.L., Martínez-Montoya J.F., Sigala-Rodríguez J.J., Esparza-Estrada C.E., Rosas-Rosas O.C., Ávila-Herrera L., Barbosa A.M. Rattlesnake (Crotalus spp.) distribution and diversity in Zacatecas, Mexico. ZooKeys. 2020;1005:103. doi: 10.3897/zookeys.1005.56964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lek S., Guégan J.-F. Artificial neural networks as a tool in ecological modelling, an introduction. Ecol. Model. 1999;120:65–73. [Google Scholar]
- León-Núñez L.J., Camero-Ramos G., Gutiérrez J.M. Epidemiology of snakebites in Colombia (2008-2016) Revista de Salud Pública. 2020;22:1–8. doi: 10.15446/rsap.V22n3.87005. [DOI] [PubMed] [Google Scholar]
- Leonelli S. What difference does quantity make? On the epistemology of Big Data in biology. Big data & society. 2014;1 doi: 10.1177/2053951714534395. 2053951714534395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levy O., Dayan T., Porter W.P., Kronfeld‐Schor N. Time and ecological resilience: can diurnal animals compensate for climate change by shifting to nocturnal activity? Ecol. Monogr. 2019;89 [Google Scholar]
- Leynaud G.C., Reati G.J. Identificación de las zonas de riesgo ofídico en Córdoba, Argentina, mediante el programa SIGEpi. Rev. Panam. Salud Públic. 2009;26:64–69. doi: 10.1590/s1020-49892009000700010. [DOI] [PubMed] [Google Scholar]
- Li X., Chen G., Liu X., Liang X., Wang S., Chen Y., Pei F., Xu X. A new global land-use and land-cover change product at a 1-km resolution for 2010 to 2100 based on human–environment interactions. Ann. Assoc. Am. Geogr. 2017;107:1040–1059. [Google Scholar]
- Licht P. Environmental physiology of reptilian breeding cycles: role of temperature. Gen. Comp. Endocrinol. 1972;3:477–488. [Google Scholar]
- Lindgren F., Rue H. Bayesian spatial modelling with R-INLA. J. Stat. Software. 2015;63:1–25. [Google Scholar]
- Lindström T., Phillips B.L., Brown G.P., Shine R. Identifying the time scale of synchronous movement: a study on tropical snakes. Movement ecology. 2015;3:1–9. doi: 10.1186/s40462-015-0038-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H. Department of Mathematics and Statistics University of Minnesota Duluth: Duluth, MN; USA: 2008. Generalized Additive Model. [Google Scholar]
- Lloyd-Smith J.O., Cross P.C., Briggs C.J., Daugherty M., Getz W.M., Latto J., Sanchez M.S., Smith A.B., Swei A. Should we expect population thresholds for wildlife disease? Trends Ecol. Evol. 2005;20:511–519. doi: 10.1016/j.tree.2005.07.004. [DOI] [PubMed] [Google Scholar]
- Long H., Mojo D., Fu C., Wang G., Kanga E., Oduor A.M., Zhang L. Patterns of human-wildlife conflict and management implications in Kenya: a national perspective. Hum. Dimens. Wildl. 2020;25:121–135. [Google Scholar]
- Longbottom J., Shearer F.M., Devine M., Alcoba G., Chappuis F., Weiss D.J., Ray S.E., Ray N., Warrell D.A., de Castañeda R.R. Vulnerability to snakebite envenoming: a global mapping of hotspots. Lancet. 2018;392:673–684. doi: 10.1016/S0140-6736(18)31224-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Löwenborg K., Shine R., Kärvemo S., Hagman M. Grass snakes exploit anthropogenic heat sources to overcome distributional limits imposed by oviparity. Funct. Ecol. 2010;24:1095–1102. [Google Scholar]
- Luz P.M., Struchiner C.J., Galvani A.P. Modeling transmission dynamics and control of vector-borne neglected tropical diseases. PLoS Neglected Trop. Dis. 2010;4 doi: 10.1371/journal.pntd.0000761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macartney J.M., Gregory P.T., Larsen K.W. A tabular survey of data on movements and home ranges of snakes. J. Herpetol. 1988:61–73. [Google Scholar]
- Madden F. Creating coexistence between humans and wildlife: global perspectives on local efforts to address human–wildlife conflict. Hum. Dimens. Wildl. 2004;9:247–257. [Google Scholar]
- Madsen T., Shine R. Seasonal migration of predators and prey‐‐A study of pythons and rats in tropical Australia. Ecology. 1996;77:149–156. [Google Scholar]
- Maina J., Ouma P.O., Macharia P.M., Alegana V.A., Mitto B., Fall I.S., Noor A.M., Snow R.W., Okiro E.A. A spatial database of health facilities managed by the public health sector in sub Saharan Africa. Sci. Data. 2019;6:1–8. doi: 10.1038/s41597-019-0142-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mamo A., Lemessa D., Diriba O.H., Hunde D. Pattern of crop raiding by wild large mammals and the resultant impacts vary with distances from forests in Southwest Ethiopia. Ecol. Evol. 2021 doi: 10.1002/ece3.7268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manral U., Sengupta S., Hussain S.A., Rana S., Badola R. Human wildlife conflict in India: a review of economic implication of loss and preventive measures. Indian For. 2016;142:928–940. [Google Scholar]
- Marshall R.J. A review of methods for the statistical analysis of spatial patterns of disease. J. Roy. Stat. Soc. 1991;154:421–441. [Google Scholar]
- McCullagh P. 2019. Generalized Linear Models. [Google Scholar]
- Medina-Barrios O.D., Hernández-Cuadrado É.E., Hernández Vélez D. Termobiología de Bothrops asper (Garman, 1883) en Colombia: ensayos ecofisiológicos. Rev. Invest. Vet. Perú. 2019;30:61–73. [Google Scholar]
- Meier P., Bergelund J. Field-testing the first cargo drone deliveries in the Amazon rainforest. Lima, Peru. 2017 http://werobotics.org/wp-content/uploads/2017/02 [Google Scholar]
- Meiri S., Roll U., Grenyer R., Feldman A., Novosolov M., Bauer A.M. Dryad; Dataset: 2017. Data from: the Global Distribution of Tetrapods Reveals a Need for Targeted Reptile Conservation. [DOI] [PubMed] [Google Scholar]
- Mendonça-da-Silva I., Tavares A.M., Sachett J., Sardinha J.F., Zaparolli L., Santos M.F.G., Lacerda M., Monteiro W.M. Safety and efficacy of a freeze-dried trivalent antivenom for snakebites in the Brazilian Amazon: an open randomized controlled phase IIb clinical trial. PLoS Neglected Trop. Dis. 2017;11 doi: 10.1371/journal.pntd.0006068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Messmer T.A. The emergence of human–wildlife conflict management: turning challenges into opportunities. Int. Biodeterior. Biodegrad. 2000;45:97–102. [Google Scholar]
- Midtgaard R. 2021. RepFocus - A Survey of the Reptiles of the World.https://repfocus.dk/ [Google Scholar]
- Mion G., Olive F. Réanimation tropicale; Paris: 1997. Les envenimations par vipéridés en Afrique Noire; pp. 349–366. Arnette. [Google Scholar]
- Mirza M.M.Q. Climate change and extreme weather events: can developing countries adapt? Clim. Pol. 2003;3:233–248. [Google Scholar]
- Mizsei E., Szabolcs M., Szabó L., Boros Z., Mersini K., Roussos S.A., Dimaki M., Ioannidis Y., Végvári Z., Lengyel S. Determining priority areas for an Endangered cold-adapted snake on warming mountaintops. Oryx. 2020:1–10. [Google Scholar]
- Mohapatra B., Warrell D.A., Suraweera W., Bhatia P., Dhingra N., Jotkar R.M., Rodriguez P.S., Mishra K., Whitaker R., Jha P. Snakebite mortality in India: a nationally representative mortality survey. PLoS Neglected Trop. Dis. 2011;5 doi: 10.1371/journal.pntd.0001018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molesworth A.M., Harrison R., David R., Theakston G., Lalloo D.G. Geographic Information System mapping of snakebite incidence in northern Ghana and Nigeria using environmental indicators: a preliminary study. Trans. R. Soc. Trop. Med. Hyg. 2003;97:188–192. doi: 10.1016/s0035-9203(03)90115-5. [DOI] [PubMed] [Google Scholar]
- Moreno-Rueda G., Pleguezuelos J.M., Alaminos E. Climate warming and activity period extension in the Mediterranean snake Malpolon monspessulanus. Climatic Change. 2009;92:235–242. [Google Scholar]
- Mukherjee A.K. Species-specific and geographical variation in venom composition of two major cobras in Indian subcontinent: impact on polyvalent antivenom therapy. Toxicon. 2020;188:150–158. doi: 10.1016/j.toxicon.2020.10.024. [DOI] [PubMed] [Google Scholar]
- Muñoz A.R., Jiménez‐Valverde A., Márquez A.L., Moleón M., Real R. Environmental favourability as a cost‐efficient tool to estimate carrying capacity. Divers. Distrib. 2015;21:1388–1400. [Google Scholar]
- Murray K.A., Martin G., Iwamura T. Focus on snake ecology to fight snakebite. Lancet. 2020;395:e14. doi: 10.1016/S0140-6736(19)32510-3. [DOI] [PubMed] [Google Scholar]
- Murray K.A., Olivero J., Roche B., Tiedt S., Guégan J.F. Pathogeography: leveraging the biogeography of human infectious diseases for global health management. Ecography. 2018;41:1411–1427. doi: 10.1111/ecog.03625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mylne A.Q., Pigott D.M., Longbottom J., Shearer F., Duda K.A., Messina J.P., Weiss D.J., Moyes C.L., Golding N., Hay S.I. Mapping the zoonotic niche of Lassa fever in Africa. Trans. R. Soc. Trop. Med. Hyg. 2015;109:483–492. doi: 10.1093/trstmh/trv047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Najmanová L., Adamík P. Effect of climatic change on the duration of the breeding season in three European thrushes. Hous. Theor. Soc. 2009;56:349–356. [Google Scholar]
- Neutens T. Accessibility, equity and health care: review and research directions for transport geographers. J. Transport Geogr. 2015;43:14–27. [Google Scholar]
- Newman W., Moran N., Theakston R., Warrell D., Wilkinson D. Traditional treatments for snake bite in a rural African community. Ann. Trop. Med. Parasitol. 1997;91:967–969. doi: 10.1080/00034989760392. [DOI] [PubMed] [Google Scholar]
- Nogueira C.C., Argôlo A.J., Arzamendia V., Azevedo J.A., Barbo F.E., Bérnils R.S., Bolochio B.E., Borges-Martins M., Brasil-Godinho M., Braz H. Atlas of Brazilian snakes: verified point-locality maps to mitigate the Wallacean shortfall in a megadiverse snake fauna. South Am. J. Herpetol. 2019;14:1–274. [Google Scholar]
- Nori J., Carrasco P.A., Leynaud G.C. Venomous snakes and climate change: ophidism as a dynamic problem. Climatic Change. 2014;122:67–80. [Google Scholar]
- Nyhus P.J. Human–wildlife conflict and coexistence. Annu. Rev. Environ. Resour. 2016;41:143–171. [Google Scholar]
- O'Connor B., Bojinski S., Röösli C., Schaepman M.E. Monitoring global changes in biodiversity and climate essential as ecological crisis intensifies. Ecol. Inf. 2020;55:101033. [Google Scholar]
- Ochoa C., Bolon I., Durso A.M., Ruiz de Castañeda R., Alcoba G., Babo Martins S., Chappuis F., Ray N. Assessing the increase of snakebite incidence in relationship to flooding events. J. Environ. Public Health. 2020 2020. [Google Scholar]
- Ogawa T., Nakashima K.-I., Nobuhisa I., Deshimaru M., Shimohigashi Y., Fukumaki Y., Sakaki Y., Hattori S., Ohno M. Accelerated evolution of snake venom phospholipase A2 isozymes for acquisition of diverse physiological functions. Toxicon. 1996;34:1229–1236. doi: 10.1016/s0041-0101(96)00112-2. [DOI] [PubMed] [Google Scholar]
- Oh A.M.F., Tan C.H., Ariaranee G.C., Quraishi N., Tan N.H. Venomics of Bungarus caeruleus (Indian krait): comparable venom profiles, variable immunoreactivities among specimens from Sri Lanka, India and Pakistan. J. Proteomics. 2017;164:1–18. doi: 10.1016/j.jprot.2017.04.018. [DOI] [PubMed] [Google Scholar]
- Ortiz A.M.D., Outhwaite C.L., Dalin C., Newbold T. A review of the interactions between biodiversity, agriculture, climate change, and international trade: research and policy priorities. One Earth. 2021;4:88–101. [Google Scholar]
- Paez A., Higgins C.D., Vivona S.F. Demand and level of service inflation in Floating Catchment Area (FCA) methods. PloS One. 2019;14 doi: 10.1371/journal.pone.0218773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandey D.P., Pandey G.S., Devkota K., Goode M. Public perceptions of snakes and snakebite management: implications for conservation and human health in southern Nepal. J. Ethnobiol. Ethnomed. 2016;12:1–25. doi: 10.1186/s13002-016-0092-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patiño-Barbosa A.M., Herrera-Giraldo A.C., Lozada-Riascos C.O., Paniz-Mondolfi A.E., Suárez J.A., Rodríguez-Morales A.J. Revista Panamericana de Enfermedades Infecciosas; 2019. Snakebites mapping in municipalities of the coffee triangle region in Colombia using geographic information systems (GIS) pp. 14–20. [Google Scholar]
- Patra A., Mukherjee A.K. Proteomic analysis of Sri Lanka Echis carinatus venom: immunological cross-reactivity and enzyme neutralization potency of Indian polyantivenom. J. Proteome Res. 2020;19:3022–3032. doi: 10.1021/acs.jproteome.0c00054. [DOI] [PubMed] [Google Scholar]
- Peace N. Impact of climate change on insect, pest, disease, and animal biodiversity. Int. J. Environ. Sci. Nat. Res. Rev. 2020 [Google Scholar]
- Peterson A.T. JHU Press; 2014. Mapping Disease Transmission Risk: Enriching Models Using Biogeography and Ecology. [Google Scholar]
- Phillips S.J., Anderson R.P., Schapire R.E. Maximum entropy modeling of species geographic distributions. Ecol. Model. 2006;190:231–259. [Google Scholar]
- Phillips S.J., Dudík M. Modeling of species distributions with Maxent: new extensions and a comprehensive evaluation. Ecography. 2008;31:161–175. [Google Scholar]
- Phillips S.J., Dudík M., Elith J., Graham C.H., Lehmann A., Leathwick J., Ferrier S. Sample selection bias and presence‐only distribution models: implications for background and pseudo‐absence data. Ecol. Appl. 2009;19:181–197. doi: 10.1890/07-2153.1. [DOI] [PubMed] [Google Scholar]
- Piccolo R.L., Warnken J., Chauvenet A.L.M., Castley J.G. Location biases in ecological research on Australian terrestrial reptiles. Sci. Rep. 2020;10:1–10. doi: 10.1038/s41598-020-66719-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pintor A., Graham E., Kennard M., VanDerWal J. James Cook University, Griffith University, and Australian Government National Environmental Science Program (NESP), Northern Australia Environmental Resources Hub; 2018. Expert Vetted Distribution Models and Biodiversity Hotspot Maps of Terrestrial and Freshwater Taxa of Conservation Concern in Northern Australia. dx.doi.org/10.4225/28/5a9f31e23e80b. [Google Scholar]
- Pintor A., Kennard M., Álvarez-Romero J., Hernandez S. James Cook University; Townsville: 2019. Prioritising Threatened Species and Threatening Processes across Northern Australia: User Guide for Data. [Google Scholar]
- Pintor A.F., Schwarzkopf L., Krockenberger A.K. Rapoport's Rule: do climatic variability gradients shape range extent? Ecol. Monogr. 2015;85:643–659. [Google Scholar]
- Pintor A.F., Schwarzkopf L., Krockenberger A.K. Hydroregulation in a tropical dry-skinned ectotherm. Oecologia. 2016;182:925–931. doi: 10.1007/s00442-016-3687-1. [DOI] [PubMed] [Google Scholar]
- Pintor A.F., Winter K.L., Krockenberger A.K., Seymour J.E. Venom physiology and composition in a litter of Common Death Adders (Acanthophis antarcticus) and their parents. Toxicon. 2011;57:68–75. doi: 10.1016/j.toxicon.2010.10.001. [DOI] [PubMed] [Google Scholar]
- Pla D., Sanz L., Quesada-Bernat S., Villalta M., Baal J., Chowdhury M.A.W., León G., Gutiérrez J.M., Kuch U., Calvete J.J. Phylovenomics of Daboia russelii across the Indian subcontinent. Bioactivities and comparative in vivo neutralization and in vitro third-generation antivenomics of antivenoms against venoms from India, Bangladesh and Sri Lanka. J. Proteomics. 2019;207:103443. doi: 10.1016/j.jprot.2019.103443. [DOI] [PubMed] [Google Scholar]
- Priston N., Underdown S. A simple method for calculating the likelihood of crop damage by primates: an epidemiological approach. Int. J. Pest Manag. 2009;55:51–56. [Google Scholar]
- Queiroz G.P., Pessoa L.A., Portaro F.C., Maria de Fátima D.F., Tambourgi D.V. Interspecific variation in venom composition and toxicity of Brazilian snakes from Bothrops genus. Toxicon. 2008;52:842–851. doi: 10.1016/j.toxicon.2008.10.002. [DOI] [PubMed] [Google Scholar]
- Ray N., Ebener S. AccessMod 3.0: computing geographic coverage and accessibility to health care services using anisotropic movement of patients. Int. J. Health Geogr. 2008;7:1–17. doi: 10.1186/1476-072X-7-63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Redding D.W., Lucas T.C., Blackburn T.M., Jones K.E. Evaluating Bayesian spatial methods for modelling species distributions with clumped and restricted occurrence data. PloS One. 2017;12 doi: 10.1371/journal.pone.0187602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reisen W.K. Landscape epidemiology of vector-borne diseases. Annu. Rev. Entomol. 2010;55:461–483. doi: 10.1146/annurev-ento-112408-085419. [DOI] [PubMed] [Google Scholar]
- Reside A.E., VanDerWal J.J., Kutt A.S., Perkins G.C. Weather, not climate, defines distributions of vagile bird species. PloS One. 2010;5 doi: 10.1371/journal.pone.0013569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ridgeway G. Generalized Boosted Models: a guide to the gbm package. Update. 2007;1:2007. [Google Scholar]
- Roll U., Feldman A., Novosolov M., Allison A., Bauer A.M., Bernard R., Böhm M., Castro-Herrera F., Chirio L., Collen B. The global distribution of tetrapods reveals a need for targeted reptile conservation. Nat. Ecol. Evol. 2017;1:1677–1682. doi: 10.1038/s41559-017-0332-2. [DOI] [PubMed] [Google Scholar]
- Rosauer D., Blom M., Bourke G., Catalano S., Donnellan S., Gillespie G., Mulder E., Oliver P., Potter S., Pratt R. Phylogeography, hotspots and conservation priorities: an example from the Top End of Australia. Biol. Conserv. 2016;204:83–93. [Google Scholar]
- Rosauer D.F., Catullo R.A., VanDerWal J., Moussalli A., Moritz C. Lineage range estimation method reveals fine-scale endemism linked to Pleistocene stability in Australian rainforest herpetofauna. PloS One. 2015;10 doi: 10.1371/journal.pone.0126274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell F.E. Snake venom immunology: historical and practical considerations. J. Toxicol. Toxin Rev. 1988;7:1–82. [Google Scholar]
- Santos-Vega M., Bouma M.J., Kohli V., Pascual M. Population density, climate variables and poverty synergistically structure spatial risk in urban malaria in India. PLoS Neglected Trop. Dis. 2016;10 doi: 10.1371/journal.pntd.0005155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saravia P., Rojas E., Arce V., Guevara C., López J.C., Chaves E., Velásquez R., Rojas G., Gutiérrez J.M. Geographic and ontogenic variability in the venom of the neotropical rattlesnake Crotalus durissus: pathophysiological and therapeutic implications. Rev. Biol. Trop. 2002:337–346. [PubMed] [Google Scholar]
- Saupe E.E., Papes M., Selden P.A., Vetter R.S. Tracking a medically important spider: climate change, ecological niche modeling, and the brown recluse (Loxosceles reclusa) PloS One. 2011;6 doi: 10.1371/journal.pone.0017731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schieffelin C.D., de Queiroz A. Herpetologica; 1991. Temperature and Defense in the Common Garter Snake: Warm Snakes Are More Aggressive than Cold Snakes; pp. 230–237. [Google Scholar]
- Schneider M.C., Min K.-d., Hamrick P.N., Montebello L.R., Ranieri T.M., Mardini L., Camara V.M., Luiz R.R., Liese B., Vuckovic M. Overview of snakebite in Brazil: possible drivers and a tool for risk mapping. PLoS Neglected Trop. Dis. 2021;15 doi: 10.1371/journal.pntd.0009044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seek . 2021. Seek by iNaturalist.https://www.inaturalist.org/pages/seek_app 15/3/2021. [Google Scholar]
- Seneviratne S., Nicholls N., Easterling D., Goodess C., Kanae S., Kossin J., Luo Y., Marengo J., McInnes K., Rahimi M., Reichstein M., Sorteberg A., Vera C., Zhan X. Changes in climate extremes and their impacts on the natural physical environment. In: B C., Barros V., Stocker T.F., Qin D., Dokken D.J., Ebi K.L., Mastrandrea M.D., Mach K.J., Plattner G.-K., Allen S.K., Tignor M., Midgley P.M., editors. Managing the Risks of Extreme Events and Disasters to Advance Climate Change Adaptation [Field. 2012. pp. 109–230. A Special Report of Working Groups I and II of the Intergovernmental Panel on Climate Change (IPCC). Cambridge University Press, Cambridge, UK, and New York, NY, USA. [Google Scholar]
- Senji Laxme R., Attarde S., Khochare S., Suranse V., Martin G., Casewell N.R., Whitaker R., Sunagar K. Biogeographical venom variation in the Indian spectacled cobra (Naja naja) underscores the pressing need for pan-India efficacious snakebite therapy. PLoS Neglected Trop. Dis. 2021;15 doi: 10.1371/journal.pntd.0009150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Senji Laxme R., Khochare S., Attarde S., Suranse V., Iyer A., Casewell N.R., Whitaker R., Martin G., Sunagar K. Biogeographic venom variation in Russell's viper (Daboia russelii) and the preclinical inefficacy of antivenom therapy in snakebite hotspots. PLoS Neglected Trop. Dis. 2021;15 doi: 10.1371/journal.pntd.0009247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma P., Chettri N., Uddin K., Wangchuk K., Joshi R., Tandin T., Pandey A., Gaira K.S., Basnet K., Wangdi S. Mapping human‒wildlife conflict hotspots in a transboundary landscape, Eastern Himalaya. Global Ecol. Conserv. 2020;24 [Google Scholar]
- Sheath D., Ruiz De Castaneda R.L., Bempong N.-E., Raviglione M., Machalaba C., Pepper M.S., Vayena E., Ray N., Wernli D., Escher G. Precision global health: a roadmap for augmented action. J. Publ. Health Epidemiol. 2020;4:1–12. [Google Scholar]
- Siljander M., Kuronen T., Johansson T., Munyao M.N., Pellikka P.K. Primates on the farm–spatial patterns of human–wildlife conflict in forest-agricultural landscape mosaic in Taita Hills, Kenya. Appl. Geogr. 2020;117:102185. [Google Scholar]
- Sillero N. What does ecological modelling model? A proposed classification of ecological niche models based on their underlying methods. Ecol. Model. 2011;222:1343–1346. [Google Scholar]
- Sillero N., Campos J., Bonardi A., Corti C., Creemers R., Crochet P.-A., Isailović J.C., Denoël M., Ficetola G.F., Gonçalves J. Updated distribution and biogeography of amphibians and reptiles of Europe. Amphibia-Reptilia. 2014;35:1–31. [Google Scholar]
- Soucy J.-P.R., Slatculescu A.M., Nyiraneza C., Ogden N.H., Leighton P.A., Kerr J.T., Kulkarni M.A. High-resolution ecological niche modeling of Ixodes scapularis ticks based on passive surveillance data at the northern frontier of Lyme disease emergence in North America. Vector Borne Zoonotic Dis. 2018;18:235–242. doi: 10.1089/vbz.2017.2234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sousa L.F., Zdenek C.N., Dobson J.S., Op den Brouw B., Coimbra F.C., Gillett A., Del-Rei T.H., Chalkidis H.d.M., Sant'Anna S., Teixeira-da-Rocha M.M. Coagulotoxicity of Bothrops (lancehead pit-vipers) venoms from Brazil: differential biochemistry and antivenom efficacy resulting from prey-driven venom variation. Toxins. 2018;10:411. doi: 10.3390/toxins10100411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- South A., Dicko A., Herringer M., Macharia P.M., Maina J., Okiro E.A., Snow R.W., van der Walt A. A rapid and reproducible picture of open access health facility data in Africa to support the COVID-19 response. Wellcome Open Res. 2020;5 doi: 10.12688/wellcomeopenres.16075.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevens G.C. The latitudinal gradient in geographical range: how so many species coexist in the tropics. Am. Nat. 1989;133:240–256. [Google Scholar]
- Stockwell D. The GARP modelling system: problems and solutions to automated spatial prediction. Int. J. Geogr. Inf. Sci. 1999;13:143–158. [Google Scholar]
- Stockwell D.R., Peterson A.T. Effects of sample size on accuracy of species distribution models. Ecol. Model. 2002;148:1–13. [Google Scholar]
- Stott P. How climate change affects extreme weather events. Science. 2016;352:1517–1518. doi: 10.1126/science.aaf7271. [DOI] [PubMed] [Google Scholar]
- Strickland J.L., Smith C.F., Mason A.J., Schield D.R., Borja M., Castañeda-Gaytán G., Spencer C.L., Smith L.L., Trápaga A., Bouzid N.M. Evidence for divergent patterns of local selection driving venom variation in Mojave Rattlesnakes (Crotalus scutulatus) Sci. Rep. 2018;8:1–15. doi: 10.1038/s41598-018-35810-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sunagar K., Moran Y. The rise and fall of an evolutionary innovation: contrasting strategies of venom evolution in ancient and young animals. PLoS Genet. 2015;11 doi: 10.1371/journal.pgen.1005596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suraweera W., Warrell D., Whitaker R., Menon G., Rodrigues R., Fu S.H., Begum R., Sati P., Piyasena K., Bhatia M. Trends in snakebite deaths in India from 2000 to 2019 in a nationally representative mortality study. Elife. 2020;9 doi: 10.7554/eLife.54076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swaroop S., Grab B. Snakebite mortality in the world. Bull. World Health Organ. 1954;10:35. [PMC free article] [PubMed] [Google Scholar]
- Tarrant S., Grewal J., Yaglom H., Lawaczeck E., Venkat H. Zoonotic disease exposure risk and rabies vaccination among wildlife professionals. EcoHealth. 2020;17:74–83. doi: 10.1007/s10393-020-01469-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tatem A.J. WorldPop, open data for spatial demography. Sci. Data. 2017;4:1–4. doi: 10.1038/sdata.2017.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terribile L.C., Feitosa D.T., Pires M.G., de Almeida P.C.R., de Oliveira G., Diniz-Filho J.A.F., Silva N.J.d., Jr. Reducing Wallacean shortfalls for the coralsnakes of the Micrurus lemniscatus species complex: present and future distributions under a changing climate. PloS One. 2018;13 doi: 10.1371/journal.pone.0205164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Treves A., Wallace R.B., Naughton-Treves L., Morales A. Co-managing human–wildlife conflicts: a review. Hum. Dimens. Wildl. 2006;11:383–396. [Google Scholar]
- Urbina-Cardona J.N., Londoño-Murcia M.C., García-Ávila D.G. Spatio-temporal dymanics of snake diversity in four habitats with different degrees of anthropogenic disturbance in the Gorgona Island National Natural Park in the Colombian Pacific. Caldasia. 2008;30:479–493. [Google Scholar]
- VanDerWal J., Shoo L.P., Johnson C.N., Williams S.E. Abundance and the environmental niche: environmental suitability estimated from niche models predicts the upper limit of local abundance. Am. Nat. 2009;174:282–291. doi: 10.1086/600087. [DOI] [PubMed] [Google Scholar]
- Vasudev D., Fletcher R.J., Jr., Goswami V.R., Krishnadas M. From dispersal constraints to landscape connectivity: lessons from species distribution modeling. Ecography. 2015;38:967–978. [Google Scholar]
- Veldkamp A., Lambin E.F. Elsevier; 2001. Predicting Land-Use Change. [Google Scholar]
- VertNet VertNet: distributed databases with backbone. 2021. http://vertnet.org/about/about.html accessed 15/3/2021.
- Warrell D.A. Symposia of the Zoological Society of London; London: 1997. 14 Geographical and Intraspecies Variation in the Clinical Manifestations of Envenoming by Snakes; pp. 189–204. The Society. 1960-1999. [Google Scholar]
- Weber M.M., Stevens R.D., Diniz‐Filho J.A.F., Grelle C.E.V. Is there a correlation between abundance and environmental suitability derived from ecological niche modelling? A meta‐analysis. Ecography. 2017;40:817–828. [Google Scholar]
- Western, G., Macdonald, D.W., Loveridge, A.J., Dickman, A.J., Tyrrell, P., Russell, S., Understanding the dynamics of lion attacks on humans and livestock in southern Maasailand, Kenya. Oryx, 1-8.
- WHO . first ed. 2010. Guidelines for the Production, Control and Regulation of Snake Antivenom Immunoglobulins.http://www.who.int/bloodproducts/snake_antivenoms/SnakeAntivenomGuideline.pdf [DOI] [PubMed] [Google Scholar]
- Who . 2010. Venomous Snakes Distribution and Species Risk Categories.https://apps.who.int/bloodproducts/snakeantivenoms/database/ [Google Scholar]
- Who . 2017. Report of the Tenth Meeting of the WHO Strategic and Technical Advisory Group for Neglected Tropical Diseases.https://www.who.int/neglected_diseases/NTD_STAG_report_2017.pdf [Google Scholar]
- Who . 2018. Annex 5 to Guidelines for the Production, Control and Regulation of Snake Antivenom Immunoglobulins.https://www.who.int/bloodproducts/snake_antivenoms/snakeantivenomguide/en/) accesse 15/3/2021 [DOI] [PubMed] [Google Scholar]
- Who . WHO Press; 2019. Snakebite Envenoming - A Strategy for Prevention and Control.https://apps.who.int/iris/bitstream/handle/10665/324838/9789241515641-eng.pdf?ua=1 [DOI] [PubMed] [Google Scholar]
- Williams D., Gutiérrez J.M., Harrison R., Warrell D.A., White J., Winkel K.D., Gopalakrishnakone P. The global snake bite initiative: an antidote for snake bite. Lancet. 2010;375:89–91. doi: 10.1016/S0140-6736(09)61159-4. [DOI] [PubMed] [Google Scholar]
- Williams D.J., Gutiérrez J.-M., Calvete J.J., Wüster W., Ratanabanangkoon K., Paiva O., Brown N.I., Casewell N.R., Harrison R.A., Rowley P.D. Ending the drought: new strategies for improving the flow of affordable, effective antivenoms in Asia and Africa. J. Proteomics. 2011;74:1735–1767. doi: 10.1016/j.jprot.2011.05.027. [DOI] [PubMed] [Google Scholar]
- Williams K.J., Belbin L., Austin M.P., Stein J.L., Ferrier S. Which environmental variables should I use in my biodiversity model? Int. J. Geogr. Inf. Sci. 2012;26:2009–2047. [Google Scholar]
- WorldClim . 2021. WorldClim Maps, Graphs, Tables, and Data of the Global Climate.https://www.worldclim.org/ [Google Scholar]
- WorldPop . 2021. WoldPop Open Sptaial Demographic Data and Research.https://www.worldpop.org/ [Google Scholar]
- Xie H., You L., Wielgosz B., Ringler C. Estimating the potential for expanding smallholder irrigation in Sub-Saharan Africa. Agric. Water Manag. 2014;131:183–193. [Google Scholar]
- Yañez-Arenas C., Díaz-Gamboa L., Patrón-Rivero C., López-Reyes K., Chiappa-Carrara X. Estimating geographic patterns of ophidism risk in Ecuador. Neotropical Biodiversity. 2018;4:55–61. [Google Scholar]
- Yañez-Arenas C., Peterson A.T., Mokondoko P., Rojas-Soto O., Martínez-Meyer E. The use of ecological niche modeling to infer potential risk areas of snakebite in the Mexican state of Veracruz. PloS One. 2014;9 doi: 10.1371/journal.pone.0100957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yañez-Arenas C., Peterson A.T., Rodríguez-Medina K., Barve N. Mapping current and future potential snakebite risk in the new world. Climatic Change. 2016;134:697–711. [Google Scholar]
- Yates V., Lebas E., Orpiay R., Bale B. Management of snakebites by the staff of a rural clinic: the impact of providing free antivenom in a nurse-led clinic in Meserani, Tanzania. Ann. Trop. Med. Parasitol. 2010;104:439–448. doi: 10.1179/136485910X12743554760306. [DOI] [PubMed] [Google Scholar]
- Yousefi M., Ahmadi M., Nourani E., Behrooz R., Rajabizadeh M., Geniez P., Kaboli M. Upward altitudinal shifts in habitat suitability of mountain vipers since the last glacial maximum. PloS One. 2015;10 doi: 10.1371/journal.pone.0138087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yousefi M., Kafash A., Khani A., Nabati N. Applying species distribution models in public health research by predicting snakebite risk using venomous snakes' habitat suitability as an indicating factor. Sci. Rep. 2020;10:1–11. doi: 10.1038/s41598-020-74682-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zacarias D., Loyola R. Climate change impacts on the distribution of venomous snakes and snakebite risk in Mozambique. Climatic Change. 2019;152:195–207. [Google Scholar]
- Zancolli G., Calvete J.J., Cardwell M.D., Greene H.W., Hayes W.K., Hegarty M.J., Herrmann H.-W., Holycross A.T., Lannutti D.I., Mulley J.F. When one phenotype is not enough: divergent evolutionary trajectories govern venom variation in a widespread rattlesnake species. Proc. Royal Soc. B. 2019;286:20182735. doi: 10.1098/rspb.2018.2735. [DOI] [PMC free article] [PubMed] [Google Scholar]