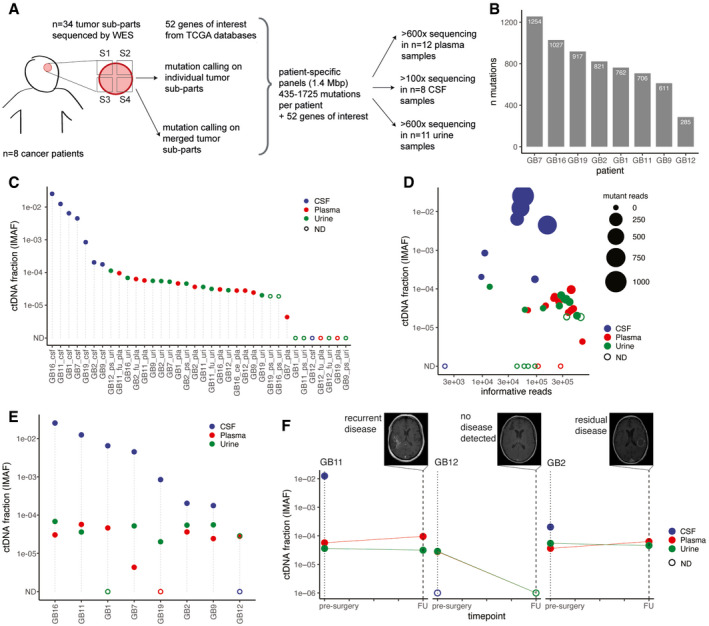
Schematic of ctDNA detection in matched CSF, plasma, and urine samples from glioma patients using INVAR (INtegration of VAriants Reads). Depth of sequencing indicated is the mean across the samples analyzed. S1 to S4 indicate tumor subparts.
Number of tumor tissue DNA mutations passing INVAR filters for the eight patients included.
Estimated ctDNA fractions for the plasma (10/12 detected), CSF (7/8 detected), and urine samples (10/16 detected) collected from 7 patients with primary glioblastoma and one patient (GB12) with anaplastic oligodendroglioma. The ctDNA fraction is expressed as IMAF (Integrated Mutant Allele Fraction). Detected cases are indicated by full circle and non‐detected cases as an open circle. ND: non‐detected.
Estimated tumor DNA DNA fraction (IMAF) in CSF, plasma, and urine depending on the number of mutant reads detected for each samples included and number of informative reads supporting the observation. Detected cases are indicated by full circle and non‐detected cases as an open circle. ND: non‐detected.
Estimated tumor DNA fractions (IMAF) in CSF, plasma, and urine for the matched samples collected at baseline pre‐surgery. Detected cases are indicated by full circle and non‐detected cases as an open circle. ND: non‐detected.
IMAF for the CSF, plasma, and urine of the patients with samples collected at 6‐month follow‐up (n = 3). Matched MRI scans are added for annotation.