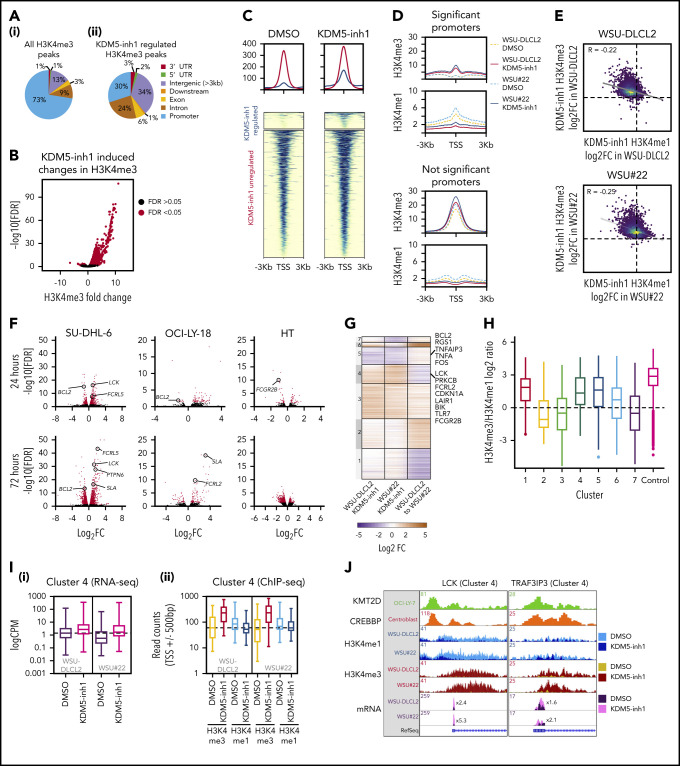
Figure 3.
Epigenetic and transcriptomic characterization of KDM5 inhibition. (A) Genomic locations of H3K4me3 peaks identified by ChIP-seq in cells treated with DMSO (i) or 1 μM KDM5-inh1 (ii) for 72 hours. (B) KDM5 inhibition induced changes in H3K4me3, with significantly changed peaks displayed in red. (C) Heatmaps of ChIP-seq data showing difference in H3K4me3 levels between promoters significantly altered (blue) or otherwise (red) in SU-DHL-6 cells treated with DMSO or 1 µM KDM5-inh1 for 72 hours. (D) Spatial plots showing distribution of H3K4me1 and H3K4me3 at promoters with significantly altered H3K4me3 by KDM5-inh1 or otherwise in WSU-DLCL2 (yellow/red) and WSU#22−/+ (light/dark blue) cells treated with DMSO (yellow/light blue) or KDM5-inh1 (red/dark blue). (E) Plots showing broad increases in H3K4me3 and reductions in H3K4me1, quantified by ChIP-seq, at the transcription start site (TSS) (±500 bp) of H3K4me3+ genes in WSU-DLCL2 and WSU#22−/+ cells treated with 1μM KDM5-inh1. The Pearson’s correlation coefficient is indicated on each plot. (F) Volcano plots indicating DE genes in SU-DHL-6, OCI-LY-18, and HT cells treated with 1 μM KDM5-inh1 for 24 and 72 hours, with significant genes highlighted in red. (G) Heatmap showing log2FC values for 897 genes that were DE by either KDM5-inh1 or KMT2D loss and clustered using K-means clustering. (H) H3K4me3 and H3K4me1 reads were counted for the promoters in each cluster and then divided (H3K4me3/H3K4me1) and log2 normalized to create a summary ratio for each. Control promoters were identified as being H3K4me3+ in WSU-DLCL2 cells but showing no alteration in mRNA expression or H3K4me3/H3K4me1 deposition in any of our analyses. (I) Boxplots showing RNA-seq logCPM values (i) and TSS read counts across ChIP-seq (ii) datasets of genes from cluster 4. (J) ChIP-seq and RNA-seq tracks, centered on LCK and TRAF3IP3, from WSU-DLCL2 and WSU#22−/+ cells treated with KDM5-inh1 for 72 hours, plus ChIP-seq tracks of KMT2D (OCI-LY-7)9 and CREBBP (centroblasts)26 binding.