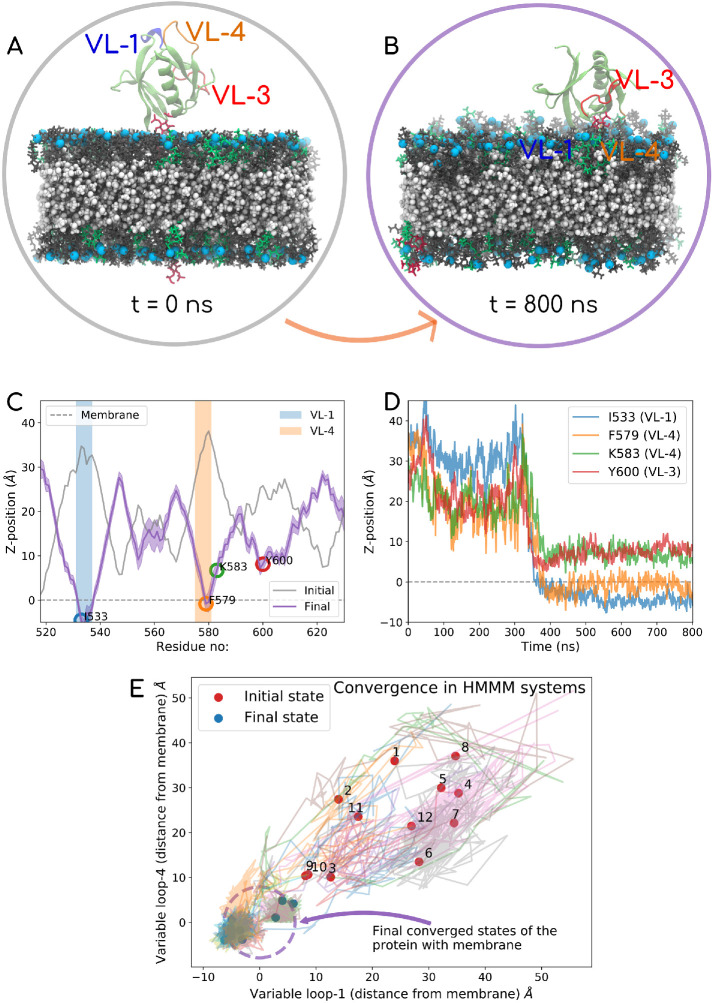
FIGURE 5:
Top panel shows the data for one of the 12 HMMM runs. In this case, the PHD is initially positioned such that all loops face away from the membrane. The PC, PS, and PIP2 mimetic lipids are shown in gray, green, and red, respectively, and phosphorus is marked in light blue. DCLE is shown in white in the core of the bilayer. The VL1 of the PHD is shown in blue, VL3 in red, and VL4in orange color. The top panel shows the initial (A) and final (B) PHD configurations. (C) Initial and final z-distance of each of the residues averaged over the last 100 ns. (D) Time evolution of signature residues on different loops. The entire 800 ns trajectory data for this system are also reported in Supplemental Movie-HMMM). In the left panel, (E) shows the convergence data for all 12 HMMM systems, where the distance from the membrane surface for the VL1 loop (I533) on the x-axis and VL4 (F579) on the y-axis is tracked with time. Almost all systems converge to a configuration with VL1 and VL4 physically engaging with the membrane. Plot akin to one on the right panel is shown for all 12 systems in Supplemental Figure S4 (i–xii)