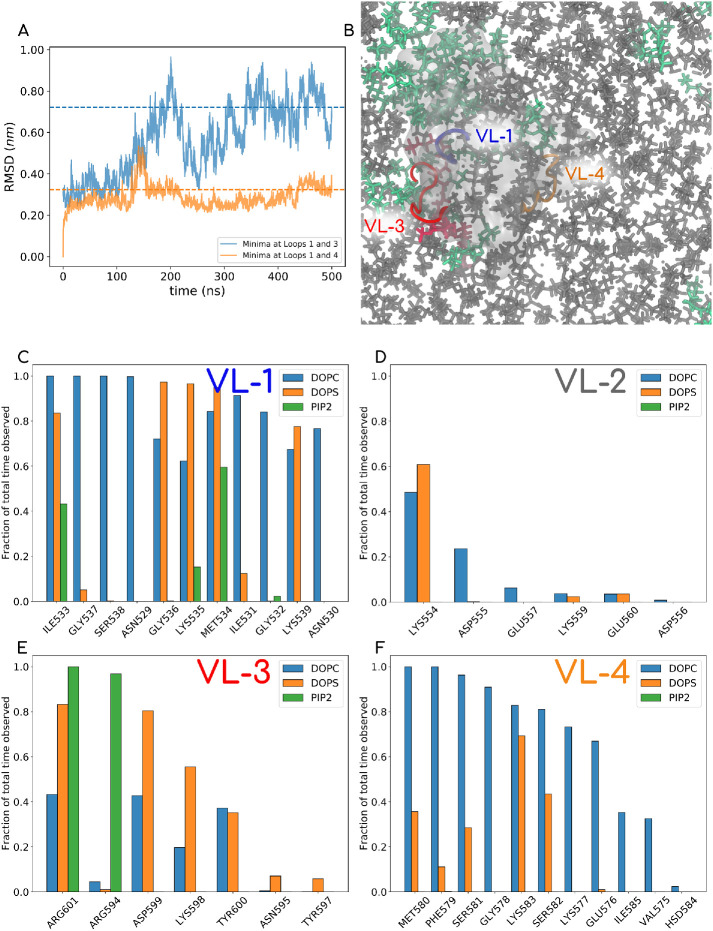
FIGURE 9:
(A) RMSD plot of AAMD runs for two systems, one started with PIP2 docking derived from a metadynamics run with IP3 ligand in VL1-VL3 and the other with IP3 ligand in the VL1-VL4 pocket. The latter shows stable convergence and was chosen for 500 ns of the MD simulation, where we explored the lipid contacts of the PHD. (B) One of the last snapshots of the MD run (water and ions not shown). PC lipids are shown in gray, PS in green, and PIP2 lipid in red. The PHD is shown in a transparent white isosurf representation, and VL1, VL3, and VL4 are shown in opaque blue, red, and orange, respectively. The full movie file of this trajectory is also reported in Supplemental Movie-AA. (C–F) Normalized contacts that different kinds of lipids make with residues on various loops.