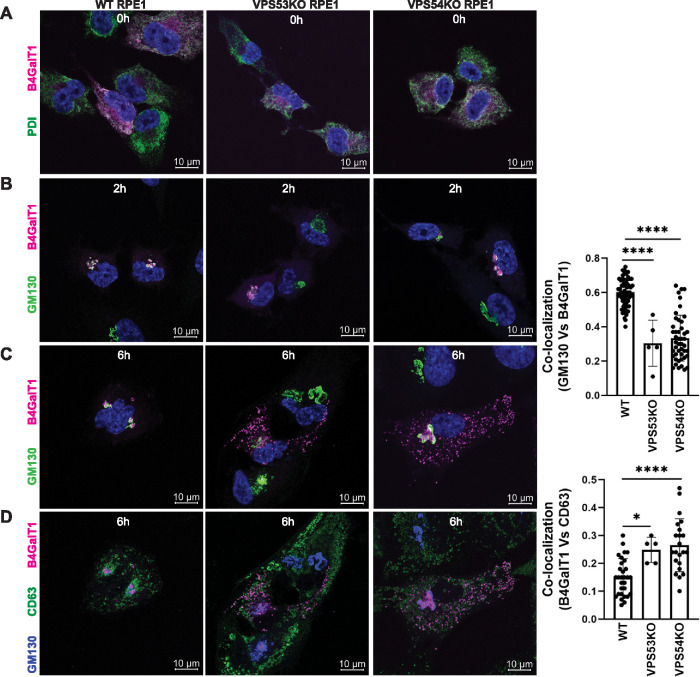
FIGURE 5:
RUSH assay reveals mislocalization of B4GalT1 to endolysosomes in GARP-KO RPE1 cells. RPE1 cells were transfected with plasmids encoding Str-KDEL_flB4GalT1-SBP-mCherry in biotin-free medium. Following overnight incubation, biotin (40 µM) and cycloheximide (50 µM) were added for 0, 2, and 6 h. (A) Colocalization of B4GalT1 with ER marker PDI at time 0. (B, C) Colocalization of B4GalT1 with Golgi protein GM130 at 2 h (B) and 6 h (C). The graph on the right side of C shows the quantification of B4GalT1 and GM130 colocalization at 6 h of biotin/cycloheximide addition. Values in the bar graph represent the mean ± SD of the colocalization between B4GalT1 and GM130 in approximately 40 cells in WT and VPS54-KOs. Colocalization in VPS53-KO cells was measured using each field in approximately 20 cells. Statistical significance was calculated using one-way ANOVA. ****P ≤ 0.0001. (D) Colocalization of B4GalT1, GM130 and the endolysosomal marker CD63 after addition of biotin/cycloheximide mix for 6 h. Quantification of the colocalization between CD63 and B4GalT1 was done by Pearson’s correlation coefficient analysis in approximately 25 cells. Statistical significance was calculated using one-way ANOVA. ****P ≤ 0.0001, *P ≤ 0.05.