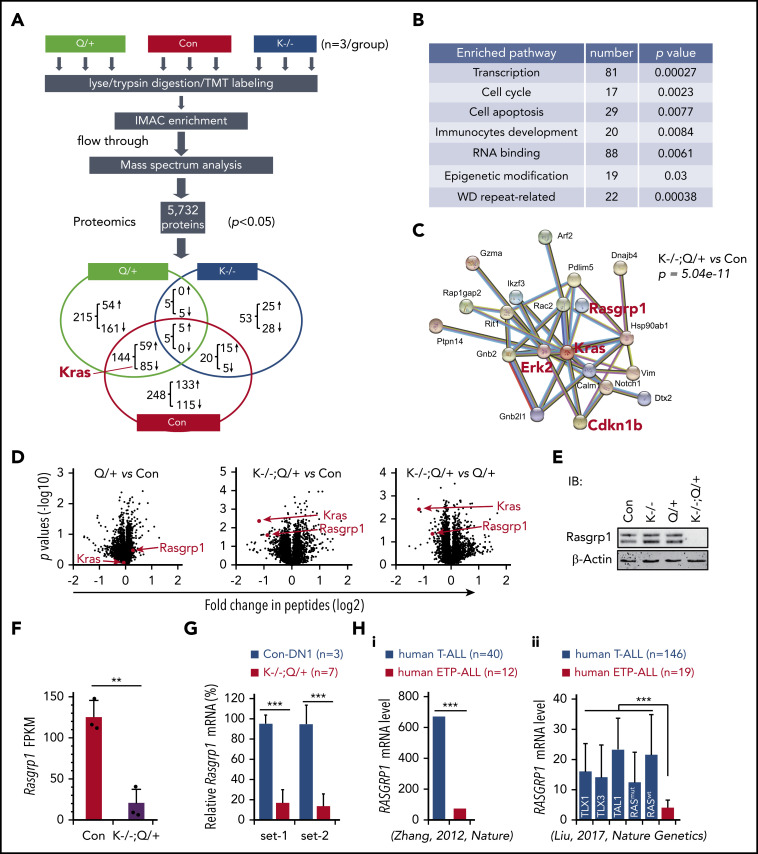
Figure 4.
Proteomics analysis identifies downregulation of Rasgrp1 in Kras−/−; NrasQ61R/+ thymocytes. Six- to 7-week-old Mx1-cre (Con), NrasQ61R/+ (Q/+), and Kras−/−; NrasQ61R/+ (K−/−; Q/+) mice were treated with pI-pC as described in "Materials and methods." Thymocytes were collected ∼4 weeks after the last pI-pC injection for analysis. (A) Flowchart of quantitative proteomics studies of thymocytes (n = 3/group). P values were calculated using a 2-tailed Student t test. Peptides with P < .05 were selected for bioinformatics analysis. (B) Peptides with P < .05 in K−/−; Q/+ vs con comparison were selected for bioinformatics analysis using the Database for Annotation, Visualization, and Integrated Discovery (DAVID) Bioinformatics Resources 6.7 software. Representative enriched pathways are shown. (C) Peptides with P < .05 and >50% changes in K−/−; Q/+ vs con comparison were selected for protein–protein interaction analysis using STRING v9.1 software. An identified Kras-centered regulatory node is shown. (D) Volcano plot analysis of total peptides in the quantitative proteomics study. The x-axis represents fold-change of total peptides (log2) in Q/+ vs control, K−/−; Q/+ vs control, and K−/−; Q/+ vs Q/+, and the y-axis represents P values (log10). (E) Western blot analysis to validate the downregulation of Rasgrp1 in K−/−; Q/+ thymic cells. (F-G) Quantification of Rasgrp1 mRNA levels using RNA-Seq in control vs K−/−; Q/+ thymocytes (F) and quantitative reverse transcription polymerase chain reaction in control thymic DN1 cells vs K−/−; Q/+ thymocytes (G). FPKM, fragments per kilobase of transcript per million mapped reads. P values were calculated with a 2-tailed Student t test. (Hi) RAGRP1 mRNA levels in human ETP-ALL patients vs general T-ALL patients. P value was cited from Zhang et al.2 (Hii) RAGRP1 mRNA levels in human ETP-ALL patients vs T-ALL patients stratified based on subtypes or RAS mutation status.34 P values were calculated with a 2-tailed Student t test. **P < .01; ***P < .001.