Figure 2.
Validation of escape mutants using yeast screening and pseudoneutralization assays
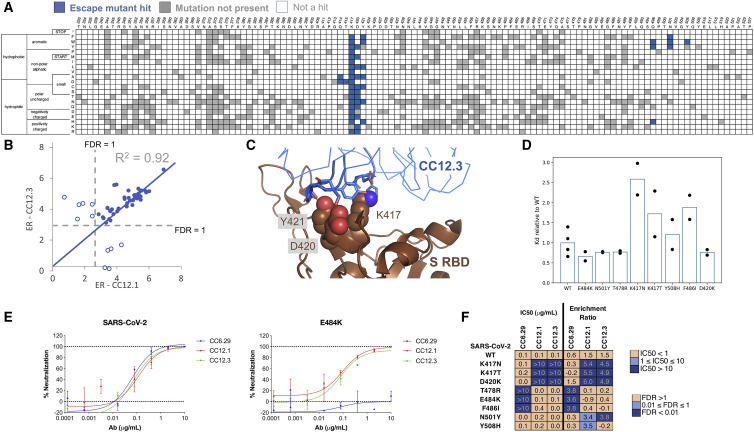
(A) Heatmap showing predicted S RBD escape mutants for CC12.3 in blue. White cells are mutations with a p value for an FDR > 1, while gray cells are mutations not present in the mutational library.
(B) Comparison of ERs for individual hits for CC12.3 versus CC12.1. Closed circles represent escape mutant hits for both nAbs, whereas open circles are escape mutant hits for only one nAb.
(C) Solved structure of nAb CC12.3 in complex with S RBD (PDB: 7KN6).
(D) Dissociation constants of single-point mutants relative to S RBD N343Q (“WT”) determined by yeast surface display titrations. Circles show the relative value for each biological replicate, and the bars represent the mean (n = 2 for each mutant; n = 4 for WT).
(E) Pseudovirus neutralization curves for CC12.1, CC12.3, and CC6.29 on SARS-CoV-2 (left) and SARS-CoV-2 E484K (right).
(F) Pseudovirus IC50 analysis for CC12.1, CC12.3, and CC6.29 on different identified mutations.
See also Figure S3.