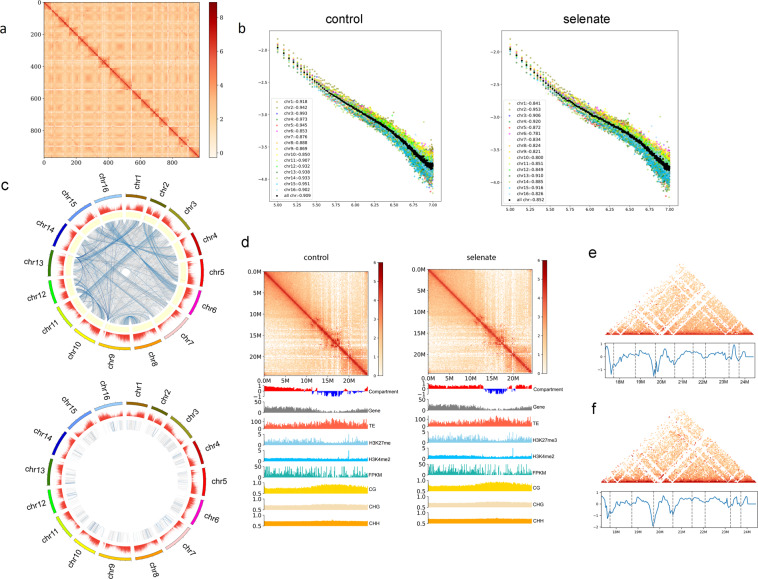
Fig. 2. Hi-C contact maps and genome-wide contact matrix of C. enshiensis.
a Hi-C interactome (400-kb bins) within and among C. enshiensis chromosomes (Chr1–Chr16). Intrachromosomal interactions were observed between the euchromatin arms of all chromosomes. The color intensity represents the frequency of contact between two 400-kb loci. Boxes along the diagonal area: interactions within the same chromosome (cis). The white rows and columns indicate bins with no valid interaction data. b Whole-genome distance-interaction frequency diagram with a 400-kb resolution between the samples. IDE values within each chromosome with or without Se supplement. Color: interaction attenuation curve of different samples; horizontal axis: relative distance between different sites on the chromosome; vertical axis: interaction frequency. c Circos plots showing genome-wide significant cis-interaction sites (upper) and trans-interaction sites (lower). d Hi-C interaction map and colocalization of the genomic composition and various epigenetic marks on Chr6. The heatmap denotes the intrachromosomal Hi-C interaction frequencies among the pairwise 100-kb bins shown on the top. The PCA eigenvectors of the A and B compartments and genomic and epigenetic feature tracks are shown below in separate 100-kb bins, including abundances of genes and TEs, different histone modifications, mRNA expression levels (normalized by FPKM), and DNA methylation (in CG, CHG, and CHH contexts). e, f Hi-C interaction matrix of a region (e chr6.116240000-24160000; f chr6.17280000-24840000) showing the TADs. Top, Hi-C interaction matrix; bottom, TAD boundaries (vertical bars) and insulation scores. The vertical axis and the blue line in the figure represent the insulation scores, and the gray line in the figure shows the TAD boundary.