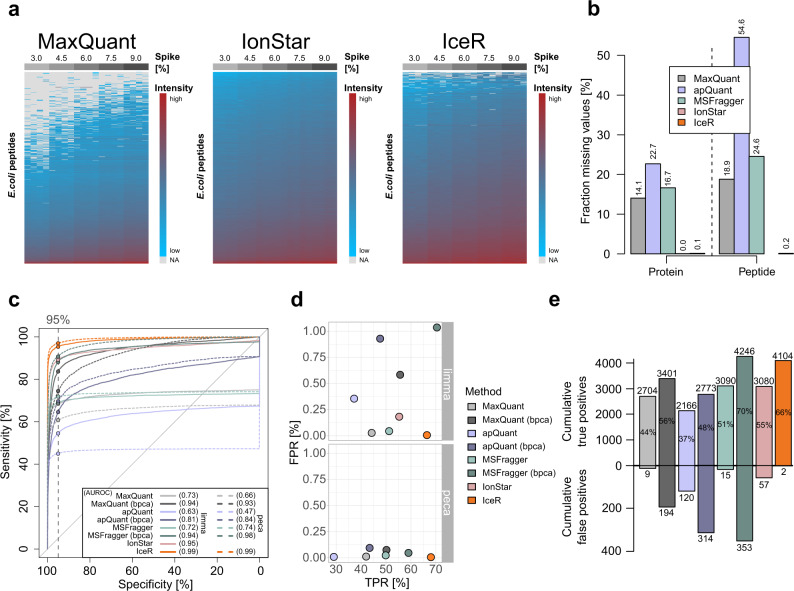
Fig. 2. IceR outperforms other label-free quantification workflows.
a Heatmap representation of quantified peptides of spiked E. coli lysate at five spike amounts into constant background (n = 4) in MaxQuant (left), IonStar (middle) and IceR (right) results. Low abundance peptides are coloured blue, high abundance peptides are coloured red, and missing values are indicated in grey. b Fraction of missing values on protein- and peptide-level in MaxQuant (grey), apQuant (purple), MSFragger (green), IonStar (pink), and IceR (orange) results. c Receiver operating characteristics (ROC) over all pairwise differential expression analyses for MaxQuant (light grey), MaxQuant with bpca imputation (dark grey), apQuant (purple), apQuant with bpca imputation (dark purple), MSFragger (green), MSFragger with bpca imputation (dark green), IonStar (pink) and IceR (orange) on protein-level using limma (solid line) and peptide-level using peca (dashed line). Area under the ROC (AUROC) per condition is indicated. Dashed vertical line and respective dots represent observed sensitivity at 95 % specificity per method. d True and false positive rates over all (10) pairwise DE analyses using limma or peca for MaxQuant (grey), MaxQuant with imputation (dark grey), apQuant (purple), apQuant with bpca imputation (dark purple), MSFragger (green), MSFragger with bpca imputation (dark green), IonStar (pink), and IceR (orange). e Cumulative true and false positives over all (10) pairwise DE analyses for MaxQuant (grey), MaxQuant with bpca imputation (dark grey), apQuant (purple), apQuant with bpca imputation (dark purple), MSFragger (green), MSFragger with bpca imputation (dark green), IonStar (pink), and IceR (orange) when using limma. True positive rates are indicated. bpca – Bayesian principal component analysis.