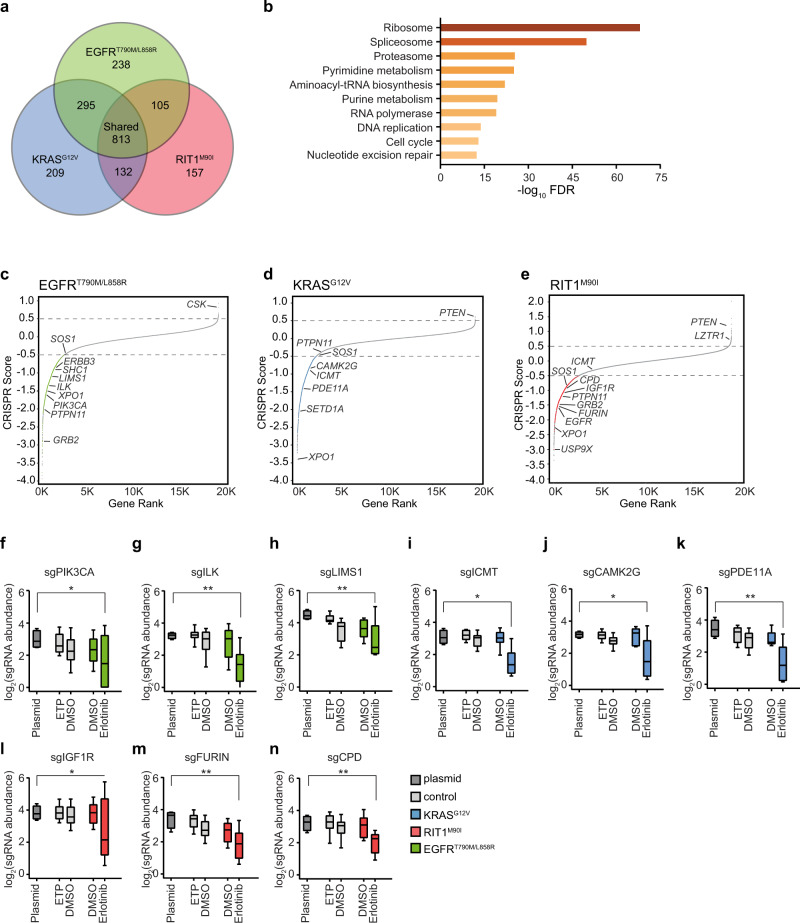
Fig. 3. Genome-wide identification of EGFRT790M/L858R, KRASG12V, and RIT1M90I oncogene-specific genetic dependencies.
a Venn Diagram showing the number of significant essential genes (CRISPR Score < 0.5 and p < 0.05) shared or specific to each isogenic PC9 cell line in erlotinib. b MSigDB overlap analysis of the enriched KEGG gene sets in the shared 813 genes from (a). c–e, Rank plots of CRISPR scores (CS) of erlotinib-treated vs. starting plasmid in (c), PC9-Cas9-EGFRT790M/L858R (d), PC9-Cas9-KRASG12V, and (e), PC9-Cas9-RIT1M90I. Key dependencies discussed in the text are labeled. Gray dashed lines mark genes with |CS| > 0.5. f–n Box plot showing the sgRNA abundance (log2 reads per million) of sgRNAs targeting each indicated gene. Abundance in the plasmid library, early time point (ETP), or after 12 population doublings in vehicle (DMSO) or erlotinib was determined by PCR and Illumina sequencing. Box plots show the median (center line), first and third quartiles (box edges), and the min and max range (whiskers) of replicates. f–h sgRNAs targeting PIK3CA (* p = 0.022), ILK (** p = 0.006), or LIMS1 (** p = 0.005) in control or PC9-Cas9-EGFRT790M/L858R cells. i–k sgRNAs targeting ICMT (* p = 0.025), CAMK2G (* p = 0.045), or PDE11A (** p = 0.004) in control or PC9-Cas9-KRASG12V. l–n sgRNAs targeting IGF1R (* p = 0.023), FURIN (** p = 0.003), or CPD (** p = 0.009) in control or PC9-Cas9-RIT1M90I cells. * p < 0.05, ** p < 0.01, calculated by one-sided permutation testing using MAGeCK. For control PC9 cells ETP and DMSO, n = 3 biological replicates. For oncogene-expressing PC9 cells DMSO and erlotinib, n = 2 biological replicates. Source data are provided as a Source Data file.