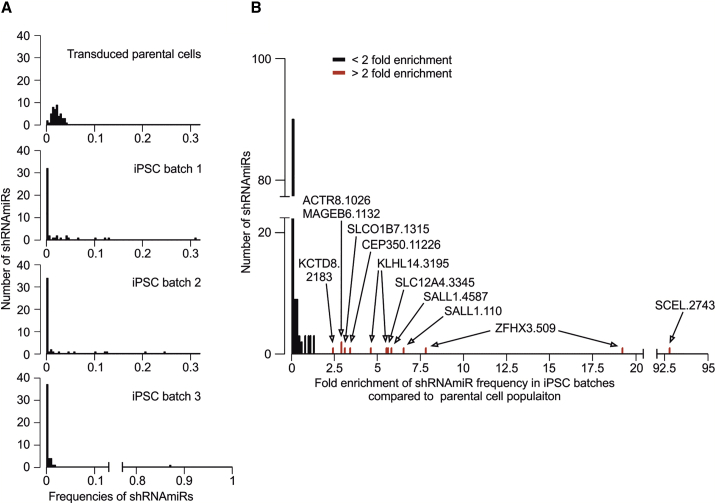
Figure 5.
Reprogramming leads to enrichment of individual shRNAmiRs directed against genes that were affected by enriched, putatively actionable GAVs
For a multiplexed shRNAmiR screen, D#2 hUVECs were transduced with a library containing shRNAmiRs against a choice of 16 genes (3 shRNAmiRs/gene) before cells were reprogrammed in 3 independent batches. The relative frequency of individual shRNAmiRs in the cell population before and after reprogramming was assessed via sequencing. (A) Frequency distribution of shRNAmiRs in transduced parental cells and the 3 iPSC batches. (B) Enrichment of shRNAmiRs displayed as fold change of shRNAmiR frequencies in iPSC batches compared to transduced starting EC population. Red marked shRNAmiRs significantly (p < 0.001) derivate from sample median (one sample, Wilcoxon signed rank test). Labels represent the names of those shRNAmiRs (Table S6) that were more than 2-fold enriched.