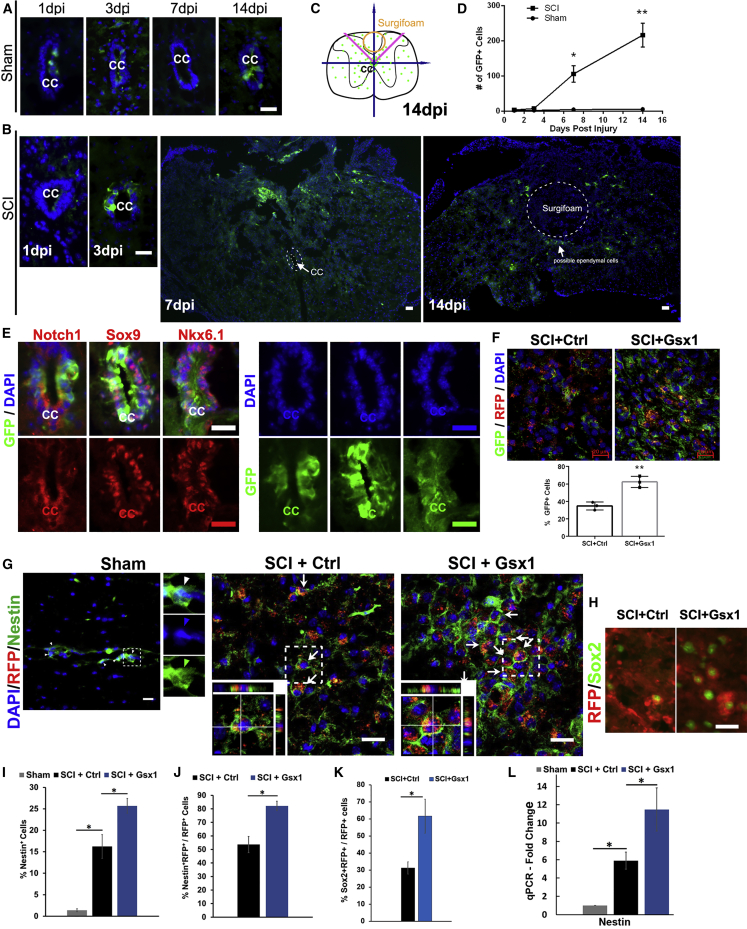
Figure 2.
Gsx1 increases the number of NSPCs after SCI
Full transection spinal cord injury (Tx) was performed on Notch1CR2-GFP transgenic mice at the T9–T10 level. (A) Fluorescence images of the cross-section through the central canal (CC) at the T9–10 level of the spinal cord from the sham groups show GFP+ cells at 1, 3, 7 and 14 DPI. Only a few GFP+ cells can be detected in the ependymal cell population lining the wall of the CC. (B) The location of GFP+ cells near the ependymal region at 1, 3, 7, and 14 DPI in the Tx animals. (C) A schematic shows the distribution of GFP+ cells. (D) Quantification of the number of GFP+ cells in the sham and SCI groups (n = 3 for all time points). (E) GFP+ ependymal cells co-labeled with Notch1, Sox9, and Nkx6.1. GFP, DAPI, and cell markers are also shown in separate channels. (F) Lentivirus injection was performed in transgenic animals with lateral hemisection SCI. Immunofluorescence analysis of the spinal cord sections shows that Gsx1 expression increased the percentage of GFP+ cells among virally transduced cells (RFP+) (n = 3). (G and H) Representative images of sagittal sections of spinal cord tissues show the expression of viral reporter RFP and the NSPC marker Nestin (G) and Sox2 (H) at 3 DPI (n = 3 for the sham group and n = 6 for the SCI+Ctrl and SCI+Gsx1). Arrows indicate Nestin+/RFP+ co-labeled cells. Images in the bottom left corner show a higher magnification z stack view of the area denoted by a dashed white box. (I–K) Quantification of all Nestin+ cells (I), Nestin+/RFP+ co-labeled cells (J), and Sox2+/RFP+ co-labeled cells (K). (L) Quantitative real-time PCR analysis shows Nestin mRNA expression at 3 DPI, normalized to the sham group; n = 4. Mean ± SEM. ∗p < 0.05 by a Students’ t test (F, J, and K) and one-way ANOVA followed by a Tukey post hoc test (D, I, and L). Scale bars, 50 μm (A–E) and 20 μm (F and G).