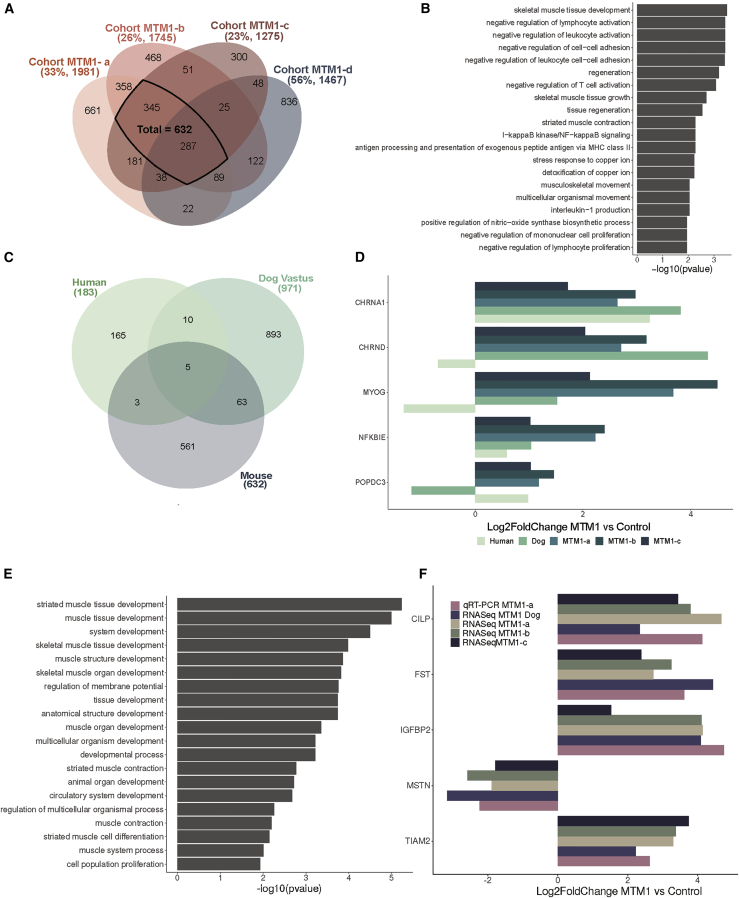
Figure 2.
MTM1-CNM signature in different species
(A) Venn diagram illustrating the shared dysregulated genes based on the Mtm1−/y versus WT comparison of four mouse cohorts. The percentages of uniquely dysregulated genes and the number of differentially expressed genes in each cohort are indicated in brackets. (B) Gene Ontology (GO) enrichment analysis of differentially expressed genes common to the four Mtm1−/y mouse cohorts. The 20 GO biological process terms with the lowest p values are displayed. (C) Venn diagram illustrating the shared dysregulated genes based on MTM1 versus control comparison in three different species: humans, mice (TA) and dogs (Vastus lateralis). (D) mRNA log2fold change expression of differentially expressed genes common to the three species. (E) GO enrichment analysis of differentially expressed genes common to mice and dogs. The 20 GO biological process terms with the lowest p value are displayed. (F) mRNA log2fold change expression of differentially expressed genes common between mice and dogs determined by RNA-seq (dogs, mice) and qRT-PCR (mice).