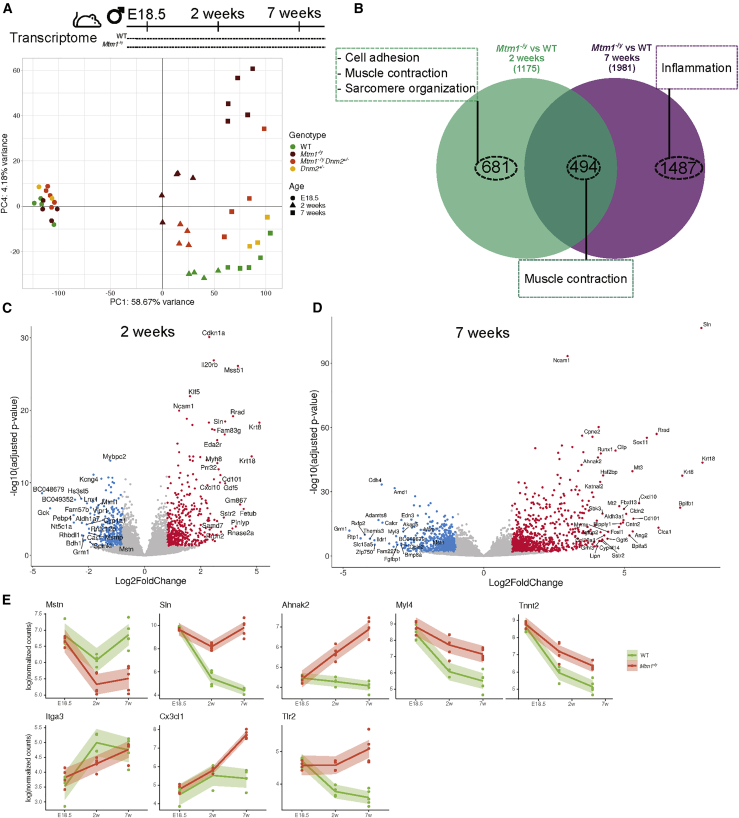
Figure 3.
Longitudinal mRNA profiling of Mtm1−/y mice
(A) Principal component analysis of RNA-seq data. The first and fourth axes are represented. Colored symbols represent genotypes, and shapes represent ages for each mouse. (B) Venn diagram illustrating the shared and specific dysregulated genes based on the Mtm1−/y versus WT comparison at 2 and 7 weeks. The most enriched GO biological processes are represented by dashed boxes. (C and D) Volcano plots representing the differentially expressed genes at (C) 2 weeks and (D) 7 weeks. Upregulated genes are in red, and downregulated genes are in blue. (E) Gene expression data (log-normalized counts) determined by RNA-seq for Mstn (muscle growth), Sln (calcium homeostasis), Ahnak2 (sarcomere organization), Myl4 and Tnnt2 (muscle contraction), Itga3 (cell adhesion), and Cxc3cl1 and Tlr2 (inflammation pathway) across time. Each dot represents an individual mouse; the shaded area represents the confidence interval at 0.95.