Abstract
The objectives of the study were to evaluate the relationship between gentamicin (GEN) and hearing loss using the Food and Drug Administration Adverse Event Reporting system (FAERS) database and elucidate the potential toxicological mechanism of GEN-induced hearing loss through a drug–gene network analysis.
Using the preferred terms and standardized queries from the Medical Dictionary for Regulatory Activities, we calculated the reporting odds ratios (RORs). We extracted GEN-associated genes (seed genes) and analyzed drug−gene interactions using the ClueGO plug-in in the Cytoscape software and the DIseAse MOdule Detection (DIAMOnD) algorithm.
The lower limit of the 95% confidence interval (CI) of the ROR for aminoglycosides (AG) antibacterials was over 1, and the ROR was 5.5 (5.1–6.0). We retrieved 17 seed genes related to GEN from the PharmGKB and Drug Gene Interaction databases. In total, 1018 human genes interacting with GEN were investigated using ClueGO. Through Molecular Complex Detection (MCODE) analysis, we identified 17 local gene clusters. The nodes and edges of the highest-ranked local gene cluster named “Cluster 1” were 30 and 433, respectively. According to the ClueGO analysis using the Kyoto Encyclopedia of Genes and Genomes (KEGG), Cluster 1 genes were highly enriched in “oxidative phosphorylation.” According to the ClueGO analysis using ClinVar, Cluster 1 genes were highly enriched in “mitochondrial diseases,” “mitochondrial complex I deficiency,” “hereditary hearing loss and deafness,” and “Leigh syndrome.” We identified 60 GEN-associated genes using the DIAMOnD algorithm. Several GEN-associated genes in the DIAMOnD algorithm were highly enriched in “PI3K-Akt signaling pathway,” “Ras signaling pathway,” “focal adhesion,” “MAPK signaling pathway,” “regulation of actin cytoskeleton,” “oxidative phosphorylation,” and “ECM-receptor interaction.”
Our analysis demonstrated an association between several AGs and hearing loss using the FAERS database. Drug−gene network analysis demonstrated that GEN may be associated with oxidative phosphorylation-associated genes and integrin genes, which may be associated with hearing loss.
Keywords: Hearing loss, Gentamicin, Aminoglycoside, Food and Drug Administration Adverse Event Reporting System, Protein−protein interaction, Oxidative phosphorylation, Integrin
Hearing loss; Gentamicin; Aminoglycoside; Food and Drug Administration Adverse Event Reporting System; Protein−protein interaction; Oxidative phosphorylation; Integrin.
1. Introduction
Hearing loss has a significant emotional and social impact on patients and a significant effect on their quality of life, owing to their inability to recognize speech, feeling of isolation, depression, cognitive impairment, anger, and difficulty in conversing (https://www.healthinaging.org/a-z-topic/hearing-loss). About 430 million individuals, which constitutes over 5% of the world's population, require rehabilitation to address their ‘disabling’ hearing loss. By 2050, approximately 2.5 billion people are predicted to have some degree of hearing loss and at least 700 million people will require hearing rehabilitation (https://www.who.int/news-room/fact-sheets/detail/deafness-and-hearing-loss). Causes of hearing loss may include genetic factors, intrauterine infections, chronic ear infections, loud sounds/noises, age-related sensorineural degeneration, and ototoxic medicines (https://www.who.int/news-room/fact-sheets/detail/deafness-and-hearing-loss). Drug classes associated with ototoxicity include antibiotics such as aminoglycosides (AGs), platinum-based anticancer drugs, loop diuretics, and nonsteroidal anti-inflammatory drugs (Altissimi et al., 2020). However, the detailed mechanism of drugs underlying hearing loss remains unclear.
AGs such as amikacin, gentamicin (GEN), neomycin, tobramycin, etc., are broad-spectrum antibiotics that are highly active against aerobic, gram-negative bacteria, such as Enterobacteriaceae and Pseudomonas. The therapeutic index of AGs is narrow, with dose-limiting ototoxicity and nephrotoxicity (Lanvers-Kaminsky et al., 2017). Since the A1555G and C1494T mutations in the 12S rRNA gene were first reported in individuals with AG-induced and non-syndromic hearing loss, AGs have been identified as a modifying factor for hearing loss, as they modulate the phenotypic manifestation of A1555G or C1494T mutations (Prezant et al., 1993; Zhao et al., 2004; Yu et al., 2014). Furthermore, the relationship between mitochondrial gene mutations and AG-induced hearing loss has been studied in detail (Lanvers-Kaminsky et al., 2017). Recently, specific genes associated with AG-induced hearing loss have also been studied. Tao and Segil (2015) showed that GEN treatment for 3 h changed the mRNA level of more than 3000 genes in the inner ear hair cells, and bioinformatic analysis of these changes highlighted several signal transduction pathways, including the Jun N-terminal kinase (JNK) and NF-κB pathways, in addition to genes involved in the stress response, apoptosis, cell cycle control, and DNA damage repair (Tao and Segil, 2015). However, this study was limited to the specific conditions of early response to GEN in hair cells. The genes and signal transduction pathways associated with GEN-induced hearing loss have not been fully investigated.
Drugs act on human body by interacting with several proteins encoded by different genes (Zhao and Iyengar, 2012). The exploration of drug–gene interactions have greatly improved our understanding of drug toxicity (Ludovini et al., 2016). In recent years, integrated analysis using data from the Food and Drug Administration (FDA) Adverse Event Reporting System (FAERS) and drug–gene interaction analysis has been proposed as a method to expand the available information on AEs (Wu et al., 2016; Lin et al., 2017).
The recent advances in genomics and biological network analyses can help elucidate the molecular mechanisms of human diseases (Barabási et al., 2011; Boyle et al., 2017). The hypothesis that disease-associated proteins tend to interact with each other in human cells is the rationale underlying network analysis. Protein interaction data are an important bioinformatics dataset used in biomedical research. Protein-coding genes associated with a disease tend to interact with each other and agglomerate in a specific network neighborhood called the disease module (Goh et al., 2007; Menche et al., 2015; Sharma et al., 2015, 2018). Molecular Complex Detection (MCODE) is a graph theoretic clustering algorithm that detects protein–protein interaction (PPI) network modules that may represent significant molecular complexes (Bader et al., 2003). Gene Ontology (GO) (Ashburner et al., 2000) annotates genes to biological/molecular terms using a hierarchical structure. To translate the network into biological insights, functional enrichment analysis for GO biological processes is used (Gene Ontology Consortium, 2015). ClueGO integrates GO terms and can compare clusters of genes to determine their functional differences (Bindea et al., 2009). The Kyoto Encyclopedia of Genes and Genomes (KEGG) is a biological systems database that integrates genomic, chemical, and systemic functional information for cells and organisms (Kanehisa et al., 2008) and assigns genes to functional pathways (Kanehisa et al., 2002). The ClinVar database in the National Center for Biotechnology Information (NCBI) archives have submitted interpretations of the clinical and functional significance of genomic variants in any region of the human genome, including that of the mitochondria (Landrum et al., 2016). The DIseAse MOdule Detection (DIAMOnD) algorithm can quantitatively identify the full disease module related to a set of known disease proteins (Ghiassian et al., 2015). By applying the DIAMOnD algorithm to the PPI dataset, modules elucidating the biological mechanisms of a disease can be identified. These functional enrichment analyses can help elucidate the potential toxicological mechanisms of GEN-induced hearing loss.
The FAERS is a spontaneous reporting system (SRS) containing data on AEs in a real-world setting voluntarily submitted by healthcare professionals, pharmaceutical companies, and patients. This database is publicly available and can be downloaded from the FDA website (http://www.fda.gov) and used in pharmacovigilance assessments. AE signals of drug-induced hearing loss (DIHL) were detected for AGs, platinum-based compounds, loop diuretics, interferons, ribavirin, papillomavirus vaccines, drugs used in erectile dysfunction, vancomycin, erythromycin, and pancuronium using the SRS, the Japanese Adverse Drug Event Report (JADER) (Tanaka et al., 2019). We also suggested the importance of carefully monitoring patients treated with AGs for DIHL, especially during the first 2 weeks (Tanaka et al., 2019). However, the sample size of the JADER database is smaller than that of the FAERS database and contains only patient data collected in Japan. Therefore, we found it worthwhile to comprehensively investigate the profiles of AG-induced deafness extracted from larger datasets.
The objectives of the study were to evaluate the relationship between AGs and hearing loss using the FAERS database. GEN is one of the most common parenteral antibacterial agents and is approved by the US FDA and other regulatory authorities around the world. GEN is often preferred for clinical use because of its low cost and reliable bactericidal activity (Jiang et al., 2017). Therefore, in this study, we elucidate the potential toxicological mechanism of GEN-induced hearing loss through a drug–gene network analysis.
2. Materials and methods
2.1. Evaluating drug-induced hearing loss
2.1.1. Data source
Data recorded in the FAERS database from January 2004 to June 2019 were obtained from the FDA website (http://www.fda.gov). The FAERS database structure complies with international safety reporting guidelines (International Council on Harmonization, E2B). We integrated the information obtained into a relational database using FileMaker Pro 13 software (FileMaker, Inc., Santa Clara, CA, USA). In compliance with the FDA's recommendation, we used the most recent case numbers to identify duplicate reports for the same patient and excluded duplicate data from the analysis (https://www.fda.gov/drugs/surveillance/questions-and-answers-fdas-adverse-event-reporting-system-faers). DrugBank (The Metabolomics Innovation Centre, Edmonton, Canada; https://www.drugbank.ca/) is a reliable drug database used as a reference in pharmacovigilance analyses (Wishart et al., 2006). Therefore, we used DrugBank (version 4.0) as a resource for batch conversion and compilation of drug names (Suzuki et al., 2015).
2.1.2. Definition of adverse events
The AEs in the JADER were defined using the Medical Dictionary for Regulatory Activities/Japanese version 19.0 (MedDRA/J, https://www.pmrj.jp/jmo/php/indexj.php). The standardized MedDRA Queries (SMQ) are groupings of MedDRA terms, ordinarily at the preferred term (PT) level that relate to a defined medical condition or area of interest. We utilized 38 PTs for DIHL that matched the SMQ for “hearing disorders” (SMQ code: 20000171) (Table S1). The SMQ for “hearing disorders” contains 50 PTs. According to our previous report, the following 12 terms that are presumably not related to DIHL were excluded: disorders associated with inflammation (acoustic neuritis [PT code: 10063162], hemotympanum [PT code: 10063013], middle ear inflammation [PT code: 10065838], myringitis [PT code: 10061302], myringitis bullous [PT code: 10028659], otosalpingitis [PT code: 10033102], and cholesterol granuloma of the middle ear [PT code: 10008649]), hearing disorders associated with hearing aids (bone-anchored hearing aid implantation [PT code: 10070723], cochlear implant [PT code: 10009830], and hearing aid therapy [PT code: 10075385]), hyperacusis (PT code: 10020559), and tinnitus (PT code: 10043882) (Tanaka et al., 2019).
2.1.3. Signal detection
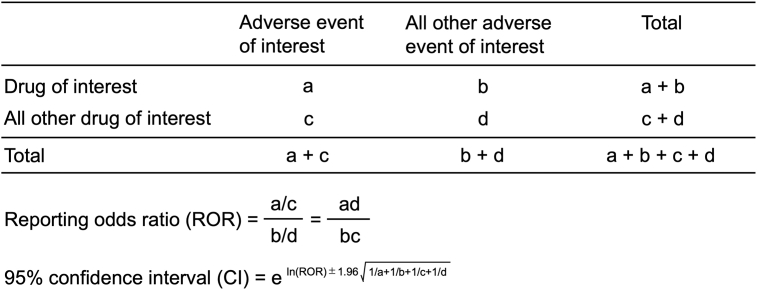
The ROR is the authorized pharmacovigilance index (van Puijenbroek et al., 2002). To detect DIHL, we calculated ROR by using a two-by-two contingency table (Fig. S1). The ROR is the ratio of the odds of reporting DIHL AEs versus all other events associated with the drug of interest compared with the reporting odds for all other drugs present in the FAERS database. A signal is defined as the lower limit of the 95% CI of the ROR being greater than one. Two or more cases were required to identify the signal (Table 1) (van Puijenbroek et al., 2002).
Table 1.
Number of reports and reporting odds ratio of drug-induced hearing loss.
| Drugs | Total (n) | Case (n) | RORb (95% CIc) |
|---|---|---|---|
| Total | 10745188 | 40073 | |
| Aminoglycoside antibacterials (ATCacode: J01G) | 32942 | 657 | 5.5 (5.1–6.0)∗ |
| streptomycin | 864 | 13 | 4.1 (2.4–7.1)∗ |
| streptoduocin | 0 | 0 | –† |
| tobramycin | 12234 | 193 | 4.3 (3.7–5.0)∗ |
| gentamicin | 9885 | 189 | 5.2 (4.5–6.0)∗ |
| kanamycin | 501 | 29 | 16.4 (11.3–23.9)∗ |
| neomycin | 3182 | 97 | 8.4 (6.9–10.3)∗ |
| amikacin | 6597 | 161 | 6.7 (5.7–7.8)∗ |
| netilmicin | 153 | 5 | 9.0 (3.7–22.0)∗ |
| sisomicin | 4 | 0 | –† |
| dibekacin | 6 | 0 | –† |
| ribostamycin | 2 | 0 | –† |
| isepamicin | 107 | 1 | –† |
| arbekacin | 248 | 0 | –† |
| bekanamycin | 1 | 0 | –† |
| plazomicin | 0 | 0 | –† |
The lower limits of 95% CI < 1.
Number of cases <2.
ATC: Anatomical Therapeutic Chemical.
ROR: reporting odds ratio.
CI: confidence interval.
2.2. ClueGO analysis
2.2.1. Human protein–protein interaction network dataset
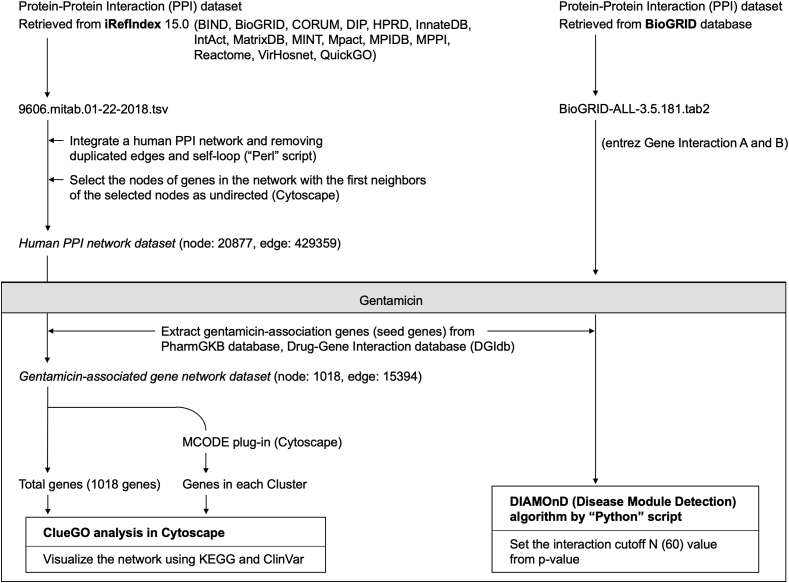
A flowchart of the data analysis is presented in Figure 1. iRefIndex provides an index of protein interactions available in several primary interaction databases including BIND (Bader et al., 2003), BioGRID (https://thebiogrid.org/), CORUM (http://mips.helmholtz-muenchen.de/corum/), DIP (https://dip.doe-mbi.ucla.edu/dip/Main.cgi), HPRD (http://www.hprd.org/), InnateDB (https://www.innatedb.com/), IntAct (https://www.ebi.ac.uk/intact/), MatrixDB (http://matrixdb.univ-lyon1.fr/), MINT (https://mint.bio.uniroma2.it/), MPact (Güldener et al., 2006), MPIDB (Goll et al., 2008), MPPI (Pagel et al., 2005), Reactome (https://reactome.org/), VirHosnet (http://virhostnet.prabi.fr/), and QuickGO (https://www.ebi.ac.uk/QuickGO/) (Razick et al., 2008). We retrieved data from the human PPI data file (9606.mitab.01-22-2018.tsv) stored in iRefIndex 15.0 (https://irefindex.vib.be/wiki/index.php/iRefIndex). We integrated a table outlining the human PPI network and removed duplicated edges and self-loops using a Perl script. Then, we imported the table to Cytoscape software (version 3.7.1), selected the nodes of genes in the network, and then selected the direction of the first neighbors of the selected nodes as “undirected.” Finally, we created a Human PPI Network dataset by compiling information from the PPI network.
Figure 1.
ClueGO and DIAMOnD analysis.
2.2.2. Extraction of seed genes from the human PPI network dataset
The drug–gene network was constructed based on drug–gene and gene–gene interactions. GEN was selected as the target drug. GEN-associated genes (seed genes) were retrieved from the PharmGKB (https://www.pharmgkb.org) and Drug Gene Interaction databases (DGIdb; https://www.dgidb.org) (Lin et al., 2017).
2.2.3. Analysis of GEN−gene interaction using ClueGO
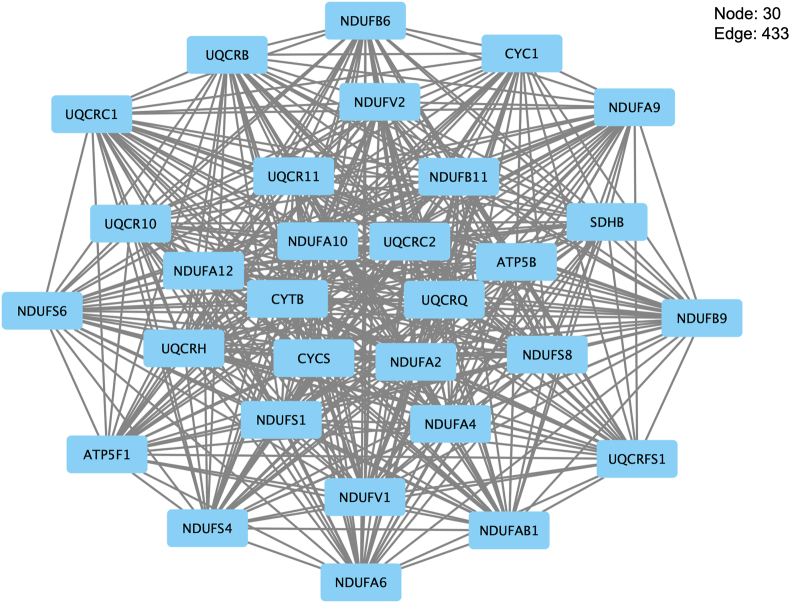
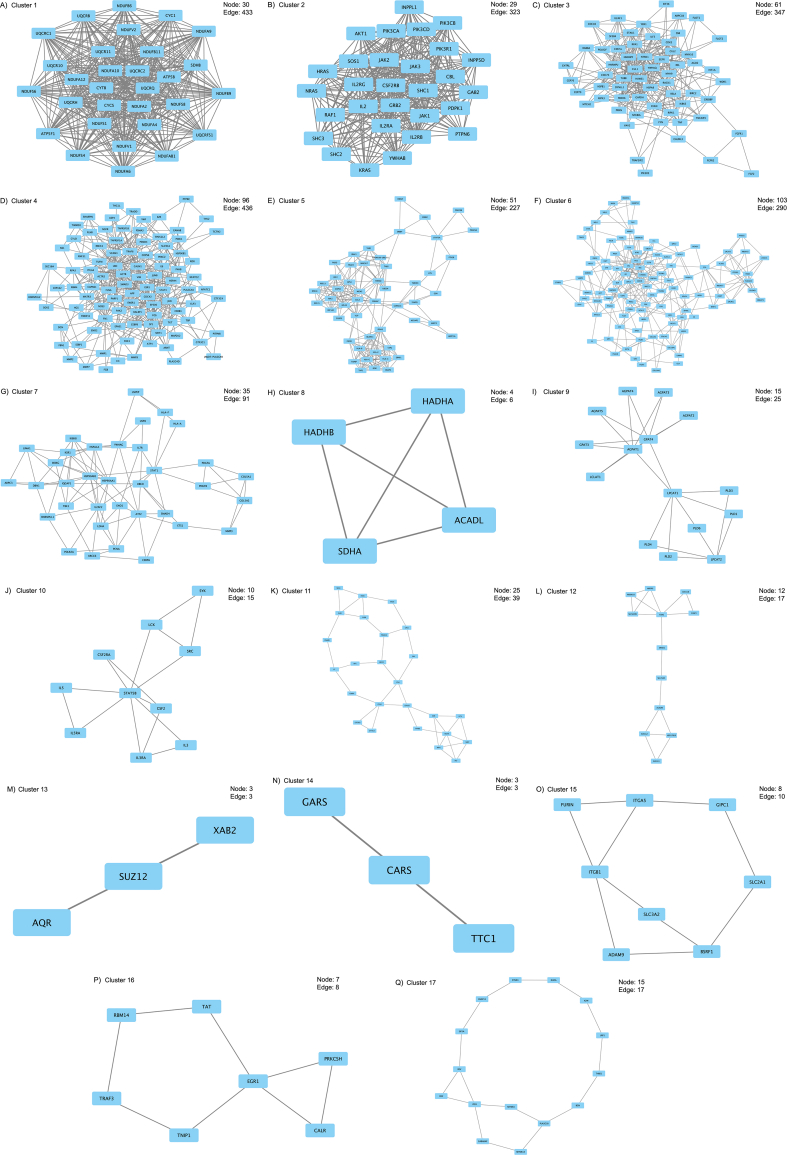
MCODE is a plug-in of Cytoscape that identifies local clusters (highly interconnect gene) that can be used to identify interesting modules (Su et al., 2014). We discovered local gene clusters from the GEN-associated gene network using MCODE (Figure 2, Fig. S2 (A–Q)). MCODE analyzes networks using scoring and finding parameters that have been optimized to produce the best results for the average user and network (https://www.baderlab.org/Software/MCODE/UsersManual). In the MCODE analysis, we selected Network Scoring Parameters and Cluster Finding Parameters by default.
Figure 2.
Gene interaction network of Cluster 1 developed using the MCODE plug-in of Cytoscape.
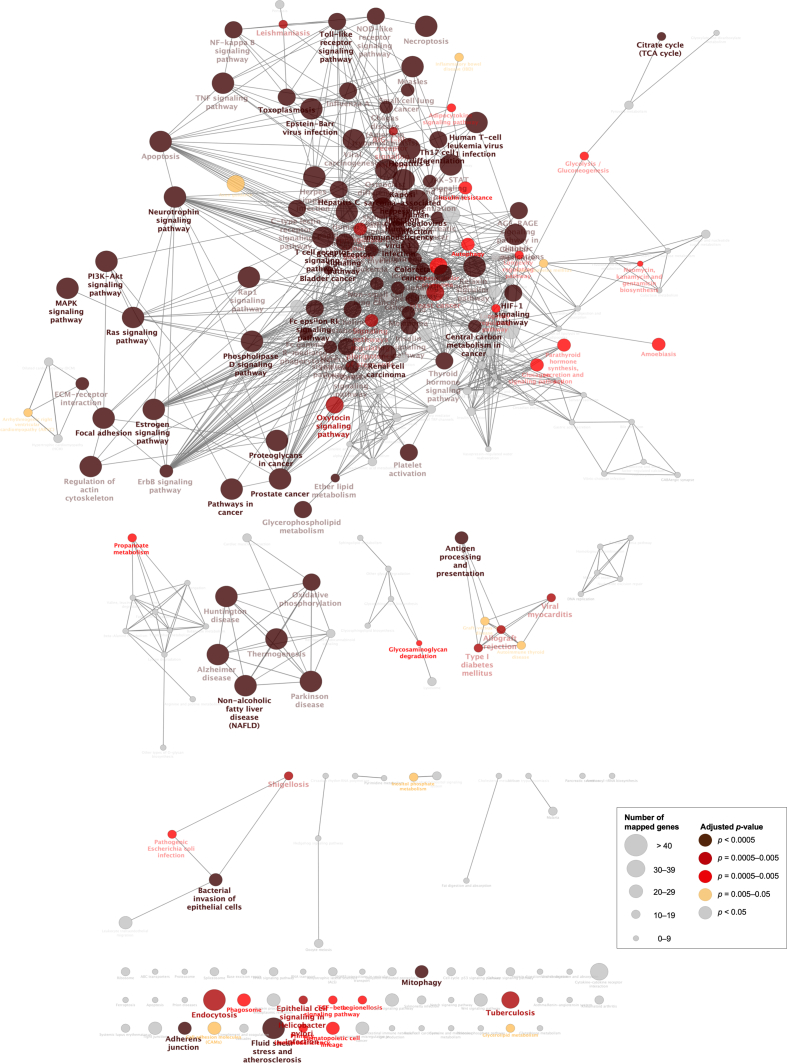
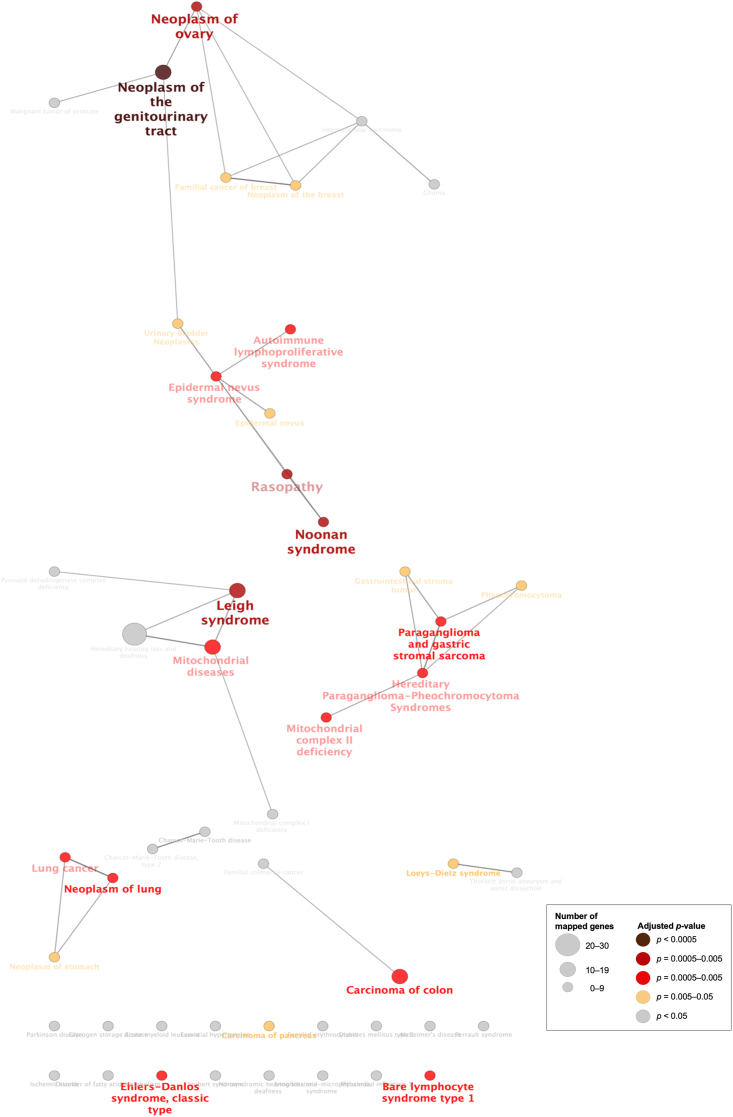
ClueGO is a plug-in of Cytoscape that integrates GO terms and creates a functionally organized GO/pathway term network. ClueGO can analyze one or two lists of genes and comprehensively visualize functionally grouped terms (Bindea et al., 2009). We selected genes from the GEN-associated gene network for the ClueGO analysis. ClueGO uses precompiled files such as GO, KEGG, and ClinVar to increase the speed of ClueGO analysis. KEGG and ClinVar (Human diseases) were used to visualize the network of molecular-level functions and clinically relevant variants in the ClueGO analysis (Figure 3, Figure 4, Figure 5, Figure 6) (Bindea et al., 2009). The raw p-values were adjusted using the Bonferroni step-down method (Table 2, Table 3, Table S2, Table S3, Table 4, Table 5).
Figure 3.
Visualization of the network of the molecular-level functions of “all” genes in the gentamicin-associated gene network. The text in the Figure 3 corresponds to the GO term in Table 2 and Table S2.
Figure 4.
Visualization of the network of the clinically relevant variants of “all” genes in the gentamicin-associated gene network. The text in the Figure 4 corresponds to the GO term in Table 3 and Table S3.
Figure 5.
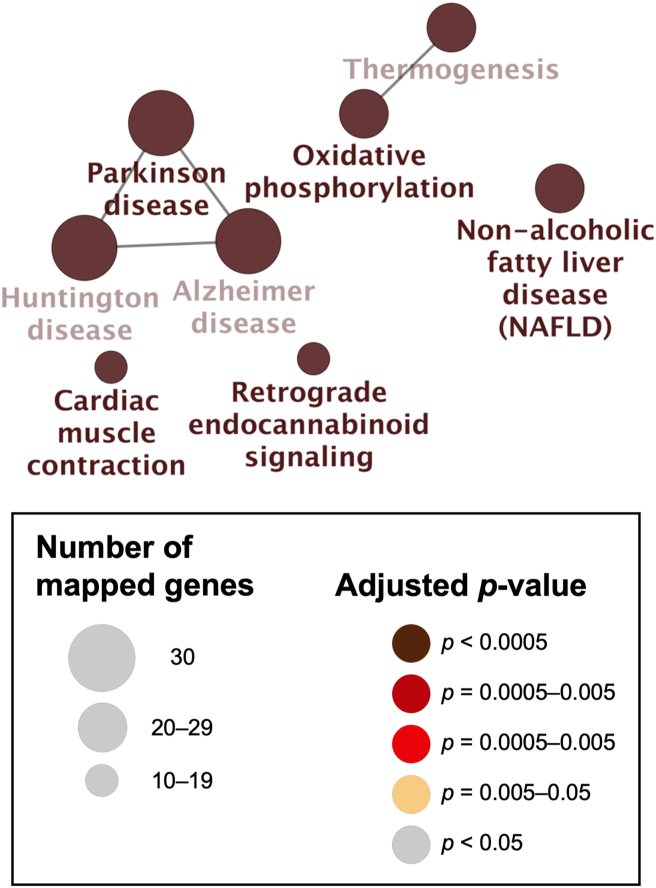
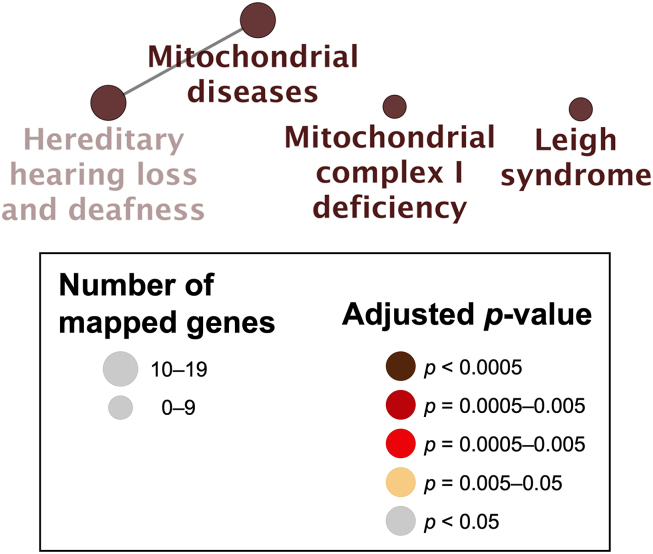
Visualization of the network of the molecular-level functions of “Cluster 1” genes in the gentamicin-associated gene network. The text in the Figure 5 corresponds to the GO term in Table 4.
Figure 6.
Visualization of the network of the clinically relevant variants of “Cluster 1” genes in the gentamicin-associated gene network. The text in the Figure 6 corresponds to the GO term in Table 5.
Table 2.
Functional annotations enriched with genes interrupted by gentamicin (partial data).
| GO ID | GO term | Adjusted p-value∗ | Associated genes found |
|---|---|---|---|
| KEGG:04151 | PI3K-Akt signaling pathway | 3.3E-20 | AKT1, ATF2, ATF4, BDNF, CDK2, CHUK, COL1A1, COL1A2, CREB1, CREB3, CREB3L1, CSF1, EGFR, ERBB2, ERBB4, FGF2, FGFR1, FGFR2, FLT1, FLT4, FN1, GRB2, GSK3B, HRAS, HSP90AA1, HSP90AB1, HSP90B1, IFNA1, IKBKB, IKBKG, IL2, IL2RA, IL2RB, IL2RG, IL3, IL3RA, IL7R, ITGA2B, ITGA3, ITGA4, ITGA5, ITGA8, ITGA9, ITGAV, ITGB1, ITGB3, ITGB5, JAK1, JAK2, JAK3, KDR, KRAS, LAMA3, MAP2K1, MAP2K2, NGFR, NOS3, NRAS, NTF3, NTF4, NTRK1, NTRK2, PDGFA, PDGFB, PDGFRA, PDGFRB, PDPK1, PGF, PIK3CA, PIK3CB, PIK3CD, PIK3R1, PIK3R5, PKN1, RAF1, RELA, RHEB, SOS1, SPP1, SYK, THBS1, TLR2, TNXB, TP53, VEGFA, VEGFB, VTN, VWF, YWHAB, YWHAE, YWHAG, YWHAQ, YWHAZ |
| KEGG:04014 | Ras signaling pathway | 4.5E-17 | AKT1, BDNF, CALM1, CALML3, CDC42, CHUK, CSF1, EGFR, ETS1, FGF2, FGFR1, FGFR2, FLT1, FLT4, GAB2, GRB2, HRAS, IKBKB, IKBKG, JMJD7-PLA2G4B, KDR, KRAS, KSR1, MAP2K1, MAP2K2, NGFR, NRAS, NTF3, NTF4, NTRK1, NTRK2, PAK1, PAK2, PDGFA, PDGFB, PDGFRA, PDGFRB, PGF, PIK3CA, PIK3CB, PIK3CD, PIK3R1, PLA2G1B, PLA2G2A, PLA2G4A, PLA2G4C, PLA2G4D, PLA2G4E, PLA2G4F, PLD1, PLD2, PRKACA, PRKACB, PTPN11, RAB5A, RAF1, RALBP1, REL, RELA, RHOA, SHC1, SHC2, SHC3, SOS1, TBK1, TIAM1, VEGFA, VEGFB |
| KEGG:04510 | Focal adhesion | 6.4E-17 | ACTB, AKT1, BIRC2, CAV1, CDC42, COL1A1, COL1A2, CTNNB1, EGFR, ERBB2, FLNA, FLNB, FLT1, FLT4, FN1, FYN, GRB2, GSK3B, HRAS, ILK, ITGA2B, ITGA3, ITGA4, ITGA5, ITGA8, ITGA9, ITGAV, ITGB1, ITGB3, ITGB5, JUN, KDR, LAMA3, MAP2K1, PAK1, PAK2, PDGFA, PDGFB, PDGFRA, PDGFRB, PDPK1, PGF, PIK3CA, PIK3CB, PIK3CD, PIK3R1, PXN, RAF1, RHOA, SHC1, SHC2, SHC3, SOS1, SPP1, SRC, THBS1, TNXB, VAV2, VEGFA, VEGFB, VTN, VWF |
| KEGG:04010 | MAPK signaling pathway | 3.1E-16 | AKT1, ATF2, ATF4, BDNF, CACNG4, CASP3, CDC42, CHUK, CSF1, EGFR, ERBB2, ERBB4, FAS, FGF2, FGFR1, FGFR2, FLNA, FLNB, FLT1, FLT4, FOS, GRB2, HRAS, HSPA1A, HSPA8, HSPB1, IKBKB, IKBKG, IRAK1, JMJD7-PLA2G4B, JUN, KDR, KRAS, MAP2K1, MAP2K2, MAP3K12, MAP3K7, NFATC1, NFATC3, NGFR, NRAS, NTF3, NTF4, NTRK1, NTRK2, PAK1, PAK2, PDGFA, PDGFB, PDGFRA, PDGFRB, PGF, PLA2G4A, PLA2G4C, PLA2G4D, PLA2G4E, PLA2G4F, PRKACA, PRKACB, RAF1, RAPGEF2, RELA, RELB, RPS6KA3, SOS1, TAB1, TAB2, TGFBR1, TGFBR2, TNF, TNFRSF1A, TP53, TRADD, TRAF2, TRAF6, VEGFA, VEGFB |
| KEGG:04722 | Neurotrophin signaling pathway | 9.9E-12 | AKT1, ATF4, BDNF, CALM1, CALML3, CDC42, GRB2, GSK3B, HRAS, IKBKB, IRAK1, JUN, KRAS, MAP2K1, MAP2K2, NFKBIA, NGFR, NRAS, NTF3, NTF4, NTRK1, NTRK2, PDPK1, PIK3CA, PIK3CB, PIK3CD, PIK3R1, PTPN11, RAF1, RELA, RHOA, RPS6KA3, SHC1, SHC2, SHC3, SORT1, SOS1, TP53, TRAF6, YWHAE |
| KEGG:04810 | Regulation of actin cytoskeleton | 1.4E-11 | ACTB, ARHGEF1, ARHGEF12, ARPC1A, ARPC1B, ARPC3, ARPC5, CDC42, EGFR, EZR, F2, FGF2, FGFR1, FGFR2, FN1, GSN, HRAS, IQGAP1, ITGA2B, ITGA3, ITGA4, ITGA5, ITGA8, ITGA9, ITGAV, ITGB1, ITGB3, ITGB5, KRAS, MAP2K1, MAP2K2, MSN, MYH10, MYH9, NCKAP1, NRAS, PAK1, PAK2, PDGFA, PDGFB, PDGFRA, PDGFRB, PIK3CA, PIK3CB, PIK3CD, PIK3R1, PXN, RAF1, RDX, RHOA, SLC9A1, SOS1, SRC, TIAM1, TMSB4X, VAV2 |
| KEGG:04210 | Apoptosis | 1.2E-09 | ACTB, AKT1, ATF4, BIRC2, CASP3, CASP8, CHUK, CSF2RB, CYCS, FADD, FAS, FOS, HRAS, IKBKB, IKBKG, IL3, IL3RA, JUN, KRAS, MAP2K1, MAP2K2, NFKBIA, NRAS, NTRK1, PARP1, PARP2, PDPK1, PIK3CA, PIK3CB, PIK3CD, PIK3R1, PRF1, RAF1, RELA, RIPK1, TNF, TNFRSF1A, TP53, TRADD, TRAF2 |
| KEGG:00190 | Oxidative phosphorylation | 2.3E-06 | ATP5F1B, ATP5PB, COX2, COX4I1, CYC1, CYTB, NDUFA10, NDUFA12, NDUFA2, NDUFA4, NDUFA6, NDUFA9, NDUFAB1, NDUFB11, NDUFB6, NDUFB9, NDUFS1, NDUFS4, NDUFS6, NDUFS8, NDUFV1, NDUFV2, SDHA, SDHB, SDHC, SDHD, UQCR10, UQCR11, UQCRB, UQCRC1, UQCRC2, UQCRFS1, UQCRH, UQCRQ |
| KEGG:04512 | ECM-receptor interaction | 8.2E-05 | CD44, COL1A1, COL1A2, DAG1, FN1, HMMR, HSPG2, ITGA2B, ITGA3, ITGA4, ITGA5, ITGA8, ITGA9, ITGAV, ITGB1, ITGB3, ITGB5, LAMA3, SPP1, THBS1, TNXB, VTN, VWF |
The raw p-values were adjusted using the Bonferroni step down method.
Table 3.
Clinically relevant annotations enriched with genes interrupted by gentamicin (partial data).
| GO ID | GO Term | Adjusted p-value∗ | Associated genes found |
|---|---|---|---|
| C0023264 | Leigh syndrome | 9.3E-04 | DLAT, DLD, NDUFA10, NDUFA12, NDUFA2, NDUFA9, NDUFS4, NDUFS8, PDHA1, SDHA |
| C0751651 | Mitochondrial diseases | 1.7E-02 | DLAT, DLD, NDUFA10, NDUFA12, NDUFA2, NDUFA9, NDUFB9, NDUFS1, NDUFS4, NDUFS6, NDUFS8, NDUFV1, NDUFV2, PDHA1, SDHA, SDHAF1, SDHD, SUCLA2, SUCLG1 |
| C1838979 | Mitochondrial complex I deficiency | 2.0E-01 | NDUFB9, NDUFS1, NDUFS4, NDUFS6, NDUFV1, NDUFV2 |
| CN043648 | Nonsyndromic hearing loss and deafness | 1.0E+00 | CLPP, MYH9, RDX |
| C0236038 | Hereditary hearing loss and deafness | 1.0E+00 | CLPP, DLAT, DLD, MYH9, NDUFA10, NDUFA12, NDUFA2, NDUFA9, NDUFB9, NDUFS1, NDUFS4, NDUFS6, NDUFS8, NDUFV1, NDUFV2, NF2, PDHA1, RDX, SDHA, SDHAF1, SDHD, SUCLA2, SUCLG1 |
The raw p-values were adjusted using the Bonferroni step down method.
Table 4.
Functional annotations enriched with Cluster 1 genes.
| GO ID | GO Term | Adjusted p-value∗ | Associated genes found |
|---|---|---|---|
| KEGG:05012 | Parkinson disease | 8.8E-53 | ATP5F1B, ATP5PB, CYC1, CYCS, CYTB, NDUFA10, NDUFA12, NDUFA2, NDUFA4, NDUFA6, NDUFA9, NDUFAB1, NDUFB11, NDUFB6, NDUFB9, NDUFS1, NDUFS4, NDUFS6, NDUFS8, NDUFV1, NDUFV2, SDHB, UQCR10, UQCR11, UQCRB, UQCRC1, UQCRC2, UQCRFS1, UQCRH, UQCRQ |
| KEGG:00190 | Oxidative phosphorylation | 1.8E-50 | ATP5F1B, ATP5PB, CYC1, CYTB, NDUFA10, NDUFA12, NDUFA2, NDUFA4, NDUFA6, NDUFA9, NDUFAB1, NDUFB11, NDUFB6, NDUFB9, NDUFS1, NDUFS4, NDUFS6, NDUFS8, NDUFV1, NDUFV2, SDHB, UQCR10, UQCR11, UQCRB, UQCRC1, UQCRC2, UQCRFS1, UQCRH, UQCRQ |
| KEGG:05010 | Alzheimer disease | 3.1E-50 | ATP5F1B, ATP5PB, CYC1, CYCS, CYTB, NDUFA10, NDUFA12, NDUFA2, NDUFA4, NDUFA6, NDUFA9, NDUFAB1, NDUFB11, NDUFB6, NDUFB9, NDUFS1, NDUFS4, NDUFS6, NDUFS8, NDUFV1, NDUFV2, SDHB, UQCR10, UQCR11, UQCRB, UQCRC1, UQCRC2, UQCRFS1, UQCRH, UQCRQ |
| KEGG:05016 | Huntington disease | 1.4E-48 | ATP5F1B, ATP5PB, CYC1, CYCS, CYTB, NDUFA10, NDUFA12, NDUFA2, NDUFA4, NDUFA6, NDUFA9, NDUFAB1, NDUFB11, NDUFB6, NDUFB9, NDUFS1, NDUFS4, NDUFS6, NDUFS8, NDUFV1, NDUFV2, SDHB, UQCR10, UQCR11, UQCRB, UQCRC1, UQCRC2, UQCRFS1, UQCRH, UQCRQ |
| KEGG:04932 | Non-alcoholic fatty liver disease (NAFLD) | 3.5E-46 | CYC1, CYCS, CYTB, NDUFA10, NDUFA12, NDUFA2, NDUFA4, NDUFA6, NDUFA9, NDUFAB1, NDUFB11, NDUFB6, NDUFB9, NDUFS1, NDUFS4, NDUFS6, NDUFS8, NDUFV1, NDUFV2, SDHB, UQCR10, UQCR11, UQCRB, UQCRC1, UQCRC2, UQCRFS1, UQCRH, UQCRQ |
| KEGG:04714 | Thermogenesis | 2.2E-43 | ATP5F1B, ATP5PB, CYC1, CYTB, NDUFA10, NDUFA12, NDUFA2, NDUFA4, NDUFA6, NDUFA9, NDUFAB1, NDUFB11, NDUFB6, NDUFB9, NDUFS1, NDUFS4, NDUFS6, NDUFS8, NDUFV1, NDUFV2, SDHB, UQCR10, UQCR11, UQCRB, UQCRC1, UQCRC2, UQCRFS1, UQCRH, UQCRQ |
| KEGG:04723 | Retrograde endocannabinoid signaling | 6.2E-20 | NDUFA10, NDUFA12, NDUFA2, NDUFA4, NDUFA6, NDUFA9, NDUFAB1, NDUFB11, NDUFB6, NDUFB9, NDUFS1, NDUFS4, NDUFS6, NDUFS8, NDUFV1, NDUFV2 |
| KEGG:04260 | Cardiac muscle contraction | 2.3E-13 | CYC1, CYTB, UQCR10, UQCR11, UQCRB, UQCRC1, UQCRC2, UQCRFS1, UQCRH, UQCRQ |
The raw p-values were adjusted using the Bonferroni step down method.
Table 5.
Clinically relevant annotations enriched with Cluster 1 genes.
| GO ID | GO Term | Adjusted p-value∗ | Associated genes found |
|---|---|---|---|
| C0751651 | Mitochondrial diseases | 7.8E-14 | NDUFA10, NDUFA12, NDUFA2, NDUFA9, NDUFB9, NDUFS1, NDUFS4, NDUFS6, NDUFS8, NDUFV1, NDUFV2 |
| C1838979 | Mitochondrial complex I deficiency | 4.6E-10 | NDUFB9, NDUFS1, NDUFS4, NDUFS6, NDUFV1, NDUFV2 |
| C0236038 | Hereditary hearing loss and deafness | 5.1E-10 | NDUFA10, NDUFA12, NDUFA2, NDUFA9, NDUFB9, NDUFS1, NDUFS4, NDUFS6, NDUFS8, NDUFV1, NDUFV2 |
| C0023264 | Leigh syndrome | 5.6E-10 | NDUFA10, NDUFA12, NDUFA2, NDUFA9, NDUFS4, NDUFS8 |
The raw p-values were adjusted using the Bonferroni step down method.
2.3. DIAMOnD analysis
The DIAMOnD algorithm presents a systematic analysis of the connectivity patterns of disease proteins and can be used to determine the predictive topological property using a Python script (Python Software Foundation, Wilmington, DW, USA). For DIAMOnD analysis, we retrieved the human PPI dataset (BioGRID-ALL-3.5.181.tab2) from the BioGRID database (https://thebiogrid.org/). To identify the GEN-associated module constructed by this algorithm, we used the human PPI dataset retrieved from the BioGRID database and the seed genes identified using PharmGKB and DGIdb. DIAMOnD is based on an iterative scheme that exploits the network topology to gradually build a drug module. In this algorithm, the p-value is determined for all genes connected to any of the s0 (s = s0) seed genes using the following formula:
| (1) |
where p (k, ki) is the hypergeometric distribution (Eq. [1]). For randomly scattered seed genes, the probability that a gene with a total of k has ks links to seed genes is given by the hypergeometric distribution:
| (2) |
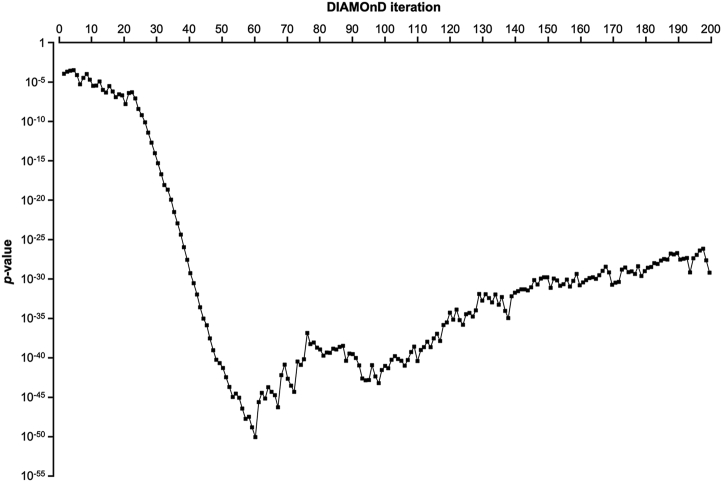
where N is the total number of genes in the network (Eq. [2]). The genes were ranked according to their respective p-values. The gene with the highest rank (i.e., lowest p-value) was added to the set of seed nodes, increasing their number from sn → sn+1 = sn + 1 (n = 0, 1, …, N−1). This operation was repeated for a fixed number of iterations N, reaching a final module size of the s0 + N genes (Ghiassian et al., 2015; Maiorino et al., 2020). We measured the p-value and obtained a curve (Table 6, Figure 7). We selected the value that yielded the lowest p-value in the curve as the iteration cutoff N (Figure 7).
Table 6.
Hypergeometric p-value of the DIAMOnD module at each iteration for gentamicin.
| Rank | DIAMOnD gene | p-value |
|---|---|---|
| 1 | DCN | 1.3E-04 |
| 2 | ABCA1 | 2.3E-04 |
| 3 | LONP2 | 3.0E-04 |
| 4 | DPP8 | 3.4E-04 |
| 5 | ATF2 | 8.4E-05 |
| 6 | AHSG | 5.5E-06 |
| 7 | S100A10 | 3.6E-05 |
| 8 | RAD18 | 1.1E-04 |
| 9 | ITGB1 | 2.1E-05 |
| 10 | ITGA3 | 3.6E-06 |
| 11 | PDIA3 | 3.8E-06 |
| 12 | ITGA2 | 1.3E-05 |
| 13 | VHL | 1.1E-06 |
| 14 | SLC3A2 | 5.1E-07 |
| 15 | HADHB | 3.2E-06 |
| 16 | CANX | 6.6E-07 |
| 17 | GPC1 | 1.3E-07 |
| 18 | CLU | 3.0E-07 |
| 19 | EGFR | 2.3E-07 |
| 20 | ITGB4 | 1.7E-08 |
| 21 | CALU | 4.5E-07 |
| 22 | HSP90B1 | 5.3E-07 |
| 23 | MYL12A | 9.1E-08 |
| 24 | COX2 | 4.2E-09 |
| 25 | COX4I1 | 6.8E-10 |
| 26 | CYC1 | 8.6E-11 |
| 27 | HADHA | 4.1E-12 |
| 28 | COX7C | 2.1E-13 |
| 29 | NDUFA9 | 9.7E-15 |
| 30 | COX7A2L | 5.3E-16 |
| 31 | UQCRC2 | 2.1E-17 |
| 32 | VDAC3 | 8.9E-19 |
| 33 | PDHB | 2.3E-19 |
| 34 | NIPSNAP1 | 1.2E-20 |
| 35 | NDUFS8 | 3.3E-22 |
| 36 | C20orf24 | 1.2E-23 |
| 37 | COA3 | 4.5E-25 |
| 38 | NDUFS1 | 1.2E-26 |
| 39 | UQCRC1 | 2.8E-28 |
| 40 | TOMM5 | 5.6E-30 |
| 41 | ATP5C1 | 2.9E-31 |
| 42 | ANKRD34C | 1.1E-32 |
| 43 | NDUFV1 | 2.6E-34 |
| 44 | UQCRQ | 9.4E-36 |
| 45 | ZNF782 | 1.3E-36 |
| 46 | NDUFS2 | 2.8E-38 |
| 47 | NDUFS3 | 8.9E-40 |
| 48 | NDUFA4 | 5.7E-41 |
| 49 | TOMM40 | 2.2E-41 |
| 50 | VDAC2 | 5.1E-42 |
| 51 | SCCPDH | 3.6E-43 |
| 52 | NDUFS7 | 2.0E-44 |
| 53 | COX7A2 | 1.1E-45 |
| 54 | NDUFS5 | 2.7E-45 |
| 55 | PHB2 | 8.5E-46 |
| 56 | MTCH2 | 3.7E-47 |
| 57 | PDHA1 | 1.7E-48 |
| 58 | CORO1C | 3.3E-48 |
| 59 | BCKDHA | 1.4E-49 |
| 60 | ATP5B | 8.5E-51 |
| 61 | TOMM22 | 2.4E-46 |
| 62 | MGST3 | 3.4E-45 |
| 63 | IMMT | 6.7E-46 |
| 64 | VDAC1 | 1.8E-44 |
| 65 | OGDH | 4.8E-45 |
| 66 | PCCB | 1.8E-45 |
| 67 | ATP6V1A | 5.0E-47 |
| 68 | NDUFA8 | 6.3E-43 |
| 69 | PHB | 1.4E-41 |
| 70 | COX6C | 2.1E-43 |
| 71 | NDUFB9 | 2.9E-44 |
| 72 | COX5A | 4.9E-45 |
| 73 | ACTB | 3.3E-41 |
| 74 | NDUFB10 | 1.3E-41 |
| 75 | ATP5O | 6.7E-41 |
| 76 | P4HA1 | 1.5E-37 |
| 77 | APOE | 5.5E-39 |
| 78 | NDUFB8 | 8.6E-39 |
| 79 | POR | 2.0E-39 |
| 80 | AGPS | 1.2E-39 |
| 81 | LRRC59 | 1.8E-40 |
| 82 | SFXN1 | 4.5E-40 |
| 83 | CYB5R3 | 4.5E-40 |
| 84 | ATP5A1 | 1.5E-39 |
| 85 | NDUFS4 | 1.1E-39 |
| 86 | UQCRH | 2.5E-39 |
| 87 | NDUFV2 | 3.4E-39 |
| 88 | NDUFA12 | 4.2E-41 |
| 89 | EMC2 | 3.4E-40 |
| 90 | HMOX2 | 3.0E-40 |
| 91 | LRPPRC | 9.5E-41 |
| 92 | NNT | 1.1E-41 |
| 93 | ASPH | 2.3E-43 |
| 94 | EMC8 | 1.4E-43 |
| 95 | ATP2A2 | 1.5E-43 |
| 96 | ATP6V1B2 | 1.2E-41 |
| 97 | RCN2 | 4.4E-43 |
| 98 | LACTB | 5.9E-44 |
| 99 | NDUFA5 | 2.9E-42 |
| 100 | RAB7A | 9.5E-42 |
| 101 | CPT1A | 4.7E-42 |
| 102 | EMC4 | 5.9E-41 |
| 103 | MINOS1 | 1.6E-40 |
| 104 | ATP5F1 | 7.3E-41 |
| 105 | ATP5H | 4.3E-41 |
| 106 | C15orf48 | 9.9E-42 |
| 107 | UQCRB | 5.2E-41 |
| 108 | SLC25A24 | 5.6E-40 |
| 109 | LONP1 | 2.6E-39 |
| 110 | TPP1 | 3.9E-41 |
| 111 | EEF2 | 9.7E-40 |
| 112 | ARMCX3 | 2.2E-39 |
| 113 | GOLT1B | 1.1E-38 |
| 114 | NDUFS6 | 2.3E-39 |
| 115 | SLC25A3 | 2.9E-38 |
| 116 | OCIAD1 | 1.1E-37 |
| 117 | AIFM1 | 1.3E-38 |
| 118 | COX8A | 1.4E-36 |
| 119 | TFAM | 3.0E-36 |
| 120 | VCP | 5.4E-35 |
| 121 | TUFM | 7.1E-36 |
| 122 | MGST1 | 1.4E-34 |
| 123 | CYB5R1 | 6.1E-36 |
| 124 | COQ9 | 1.6E-36 |
| 125 | GBAS | 3.4E-35 |
| 126 | VAPA | 4.9E-35 |
| 127 | NDUFA6 | 1.7E-35 |
| 128 | NDUFB11 | 1.0E-34 |
| 129 | HSPD1 | 1.3E-32 |
| 130 | NDUFA2 | 1.8E-33 |
| 131 | NDUFB7 | 1.2E-32 |
| 132 | ICT1 | 4.0E-33 |
| 133 | ATP5L | 1.1E-33 |
| 134 | NDUFA13 | 1.1E-32 |
| 135 | NDUFB5 | 5.6E-34 |
| 136 | AFG3L2 | 5.3E-33 |
| 137 | NDUFA11 | 9.2E-35 |
| 138 | NDUFB4 | 1.1E-35 |
| 139 | RPN1 | 6.8E-33 |
| 140 | ATP6V0D1 | 1.9E-32 |
| 141 | ATP1A1 | 2.9E-32 |
| 142 | COX14 | 5.1E-32 |
| 143 | HK1 | 4.9E-32 |
| 144 | SDHA | 3.8E-32 |
| 145 | TIMMDC1 | 9.2E-32 |
| 146 | BCKDHB | 7.4E-31 |
| 147 | SLC25A13 | 2.1E-31 |
| 148 | ATP6V1E1 | 1.2E-30 |
| 149 | ADRB2 | 1.8E-30 |
| 150 | EGLN3 | 1.7E-30 |
| 151 | STOML2 | 8.2E-32 |
| 152 | COX15 | 1.2E-30 |
| 153 | PRDX3 | 7.0E-31 |
| 154 | NDUFA3 | 1.4E-31 |
| 155 | RAB1A | 2.2E-31 |
| 156 | ATP1B1 | 8.7E-31 |
| 157 | COX6B1 | 1.2E-31 |
| 158 | LMAN1 | 6.0E-31 |
| 159 | BCAP31 | 4.6E-30 |
| 160 | NDUFA7 | 1.7E-31 |
| 161 | NDUFB1 | 3.8E-31 |
| 162 | MMGT1 | 7.4E-31 |
| 163 | MRM1 | 1.4E-30 |
| 164 | SCO1 | 1.7E-30 |
| 165 | CHCHD3 | 1.1E-30 |
| 166 | OCIAD2 | 3.1E-30 |
| 167 | USMG5 | 1.1E-29 |
| 168 | TRMT61B | 3.8E-29 |
| 169 | LETM1 | 7.2E-30 |
| 170 | ACOT9 | 1.9E-31 |
| 171 | PDK1 | 3.6E-31 |
| 172 | SLC25A20 | 4.6E-31 |
| 173 | ATP5J | 1.7E-29 |
| 174 | UNC93B1 | 3.0E-29 |
| 175 | DDOST | 7.7E-30 |
| 176 | AGR2 | 9.6E-30 |
| 177 | UQCRFS1 | 4.6E-30 |
| 178 | ZNF598 | 4.4E-29 |
| 179 | ATP1B3 | 2.5E-30 |
| 180 | P2RY6 | 1.0E-29 |
| 181 | RPN2 | 2.8E-29 |
| 182 | SAMM50 | 3.5E-29 |
| 183 | VAPB | 1.1E-28 |
| 184 | NDUFB3 | 8.8E-29 |
| 185 | NDUFV3 | 2.3E-28 |
| 186 | NDUFA10 | 3.5E-28 |
| 187 | SOAT1 | 3.1E-28 |
| 188 | GNG5 | 1.7E-27 |
| 189 | PTGES2 | 1.4E-27 |
| 190 | ESR2 | 2.0E-27 |
| 191 | CISD3 | 3.1E-28 |
| 192 | ATP5J2 | 3.7E-28 |
| 193 | ATP5D | 4.9E-28 |
| 194 | ATP5I | 7.1E-30 |
| 195 | SLC15A3 | 4.4E-28 |
| 196 | STOM | 1.2E-27 |
| 197 | MTCH1 | 4.3E-27 |
| 198 | ACAD9 | 7.4E-27 |
| 199 | NDUFAF3 | 2.4E-28 |
| 200 | NDUFAF4 | 6.8E-30 |
Figure 7.
Hypergeometric p-value of the DIAMOnD module at each iteration for gentamicin.
3. Results
3.1. Evaluating drug-induced hearing loss in the FAERS database
The FAERS database had 10 745 188 reports from January 2004 to June 2019. The number of reports for DIHL was 40 073. The top three AEs reported were hypoacusis (16 557 cases, PT code: 10048865), deafness (13 140 cases, PT code: 10011878), and deafness unilateral (3177 cases, PT code: 10048812) (Table S1). The ROR for AG antibacterials was 5.5 (5.1–6.0) and the lower limit of its 95% CI was greater than one (Table 1). The lower limits of the 95% CI of the RORs for streptomycin, tobramycin, GEN, kanamycin, neomycin, amikacin, and netilmicin were greater than one (Table 1).
3.2. Analysis of GEN−gene interaction
3.2.1. ClueGO analysis
We created the Human PPI Network dataset, and the number of nodes and edges were 20 877 and 429 350, respectively. We retrieved 17 seed genes (ANPEP, BDNF, CALR, CD44, GDNF, GGT1, HEXB, IL17A, IL2, MT-RNR, PLA2G2A, SDHB, SLC2A1, SPP1, TGM2, TNF, and VEGFA) from PharmGKB and DGIdb. We applied the retrieved seed genes to the Human PPI Network dataset and created a GEN-associated gene network. The number of nodes and edges of the GEN-associated gene network were 1018 and 15 394, respectively. In the MCODE analysis, we identified 17 local gene clusters (Figure 2, Fig. S2 (A–Q)). The nodes and edges of the highest-ranked local gene cluster named “Cluster 1” were 30 and 433, respectively.
We also evaluated the molecular-level functions and clinically relevant variants of GEN-associated genes using ClueGO for “all” genes in the GEN-associated gene network. Using KEGG, a network of molecular-level functions of GEN was visualized (Figure 3). The adjusted p-values of the “PI3K-Akt signaling pathway,” “Ras signaling pathway,” “focal adhesion,” “p38/mpk1 mitogen-activated protein kinase (MAPK) signaling pathway,” “neurotrophin signaling pathway,” “regulation of actin cytoskeleton,” “apoptosis,” “oxidative phosphorylation,” and “ECM-receptor interaction” were 3.3 × 10−20, 4.5 × 10−17, 6.4 × 10−17, 3.1 × 10−16, 9.9 × 10−12, 1.4 × 10−11, 1.2 × 10−9, 2.3 × 10−6, and 8.2 × 10−5, respectively (Table 2, Table S2). Using ClinVar, a network of clinically relevant variants of GEN was visualized (Figure 4). The adjusted p-values of the “Leigh syndrome” and “mitochondrial diseases” were 9.3 × 10−4 and 1.7 × 10−2, respectively (Table 3, Table S3).
Next, the genes in “Cluster 1” were highly enriched in “oxidative phosphorylation” (adjusted p-value = 1.8 × 10−50, 96.7% [29/30]) based on the KEGG analysis (Figure 2, Figure 5, Table 4). Several genes in Cluster 1 were highly enriched in “mitochondrial diseases” (adjusted p-value = 7.8 × 10−14, 36.7% [11/30]), “mitochondrial complex I deficiency” (adjusted p-value = 4.6 × 10−10, 20.0% [6/30]), “hereditary hearing loss and deafness” (adjusted p-value = 5.1 × 10−10, 36.7% [11/30]), and “Leigh syndrome” (adjusted p-value = 5.6 × 10−10, 20.0% [6/30]) using ClinVar (Figure 2, Figure 6, Table 5).
3.2.2. DIAMOnD analysis
Using the DIAMOnD algorithm, we measured the p-value and obtained a curve (Table 6, Figure 7). The lowest p-value in the curve was 8.5 × 10−51, and we selected the iterations cutoff N = 60 (Figure 7). Therefore, the final number of GEN-associated genes according to the DIAMOnD algorithm was 60. Some of these genes were highly enriched in “PI3K-Akt signaling pathway” (ATF2, ITGB1, ITGA3, EGFR, and HSP90B1), “Ras signaling pathway” (EGFR), “focal adhesion” (ITGB1, ITGA3, and EGFR), “MAPK signaling pathway” (ATF2 and EGFR), “regulation of actin cytoskeleton” (ITGB1, ITGA3, and EGFR), “oxidative phosphorylation” (COX2, COX4l1, CYC1, NDUFA9, UQCRC2, NDUFS8, NDUFS1, UQCRC1, NDUFV1, UQCRQ, and NDUFA4), and “ECM-receptor interaction” (ITGB1 and ITGA3) (Table 2, Table S2).
4. Discussion
4.1. ROR signals in the FAERS database
A greater percentage of patients who developed ototoxicity had high trough levels of AGs, and were older (Gatell et al., 1987). The descending order of the ototoxicity of AGs is thought to be neomycin, GEN, kanamycin, tobramycin, amikacin, and netilmicin with amikacin and netilmicin having the lowest ototoxicity (Xie et al., 2011). Our results show that AE signals for DIHL were detected for several AGs in the FAERS database (Table 1), which is corroborated by our previous report using the JADER database (Tanaka et al., 2019). Therefore, these AGs may be associated with DIHL.
4.2. GEN-induced hearing loss and related genes
4.2.1. Association between GEN and oxidative phosphorylation-associated genes
Since the A1555G and C1494T mutations in the 12S rRNA gene were first reported in families with AG-induced and non-syndromic hearing loss, AGs have been identified as a modifying factor for hearing loss as they modulate the phenotypic manifestation of A1555G or C1494T mutations (Prezant et al., 1993; Zhao et al., 2004; Yu et al., 2014). The C1494T mutation is expected to form a novel U1494-1555A base pair, which is in the same position as the C1494-1555G pair created by the deafness-associated A1555G mutation, at the highly conserved A site of 12S rRNA (Zhao et al., 2004). Sequence alterations that correspond to the deafness-associated mitochondrial mutations A1555G and C1494T increased drug-binding and rendered the ribosomal decoding site hypersusceptible to AG-induced mistranslation and inhibition of protein synthesis (Hobbie et al., 2008). AGs not only decrease mitochondrial ATP synthesis, but also induce excessive reactive oxygen species (ROS) production, which may trigger multiple forms of cell death via the JNK/MAPK or BCL2 pathways (Wang et al., 2003; Coffin et al., 2013; Yu et al., 2014). Mitochondrial calcium can stimulate oxidative phosphorylation, promoting ROS generation from respiratory complexes I and III during AG-induced hair cell death (Feissner et al., 2009; Esterberg et al., 2016; Desa et al., 2018). The A1555G mutation increases 12S rRNA hypermethylation, which causes ROS-dependent activation of AMP kinase and the proapoptotic nuclear transcription factor E2F1, and then induces apoptosis in the stria vascularis and spiral ganglion neurons (SGNs) of the inner ear and eventually, hearing loss (Raimundo et al., 2012). Therefore, AGs and mitochondrial gene mutations may increase ROS and induce cell death, leading to hearing loss.
In the ClueGO analysis using KEGG and ClinVar, “oxidative phosphorylation,” “Leigh syndrome,” and “mitochondrial diseases” were associated with GEN (Figure 3, Figure 4, Table 2, Table 3, Table S2, Table S3). Some GEN-associated genes in the DIAMOnD algorithm were highly enriched in “oxidative phosphorylation” (COX2, COX4l1, CYC1, NDUFA9, UQCRC2, NDUFS8, NDUFS1, UQCRC1, NDUFV1, UQCRQ, and NDUFA4) (Figure 3, Figure 7, Table 2, Table 6, Table S2). In the ClueGO analysis using KEGG and ClinVar, some Cluster 1 genes were highly enriched in “oxidative phosphorylation,” “mitochondrial diseases,” “mitochondrial complex I deficiency,” “hereditary hearing loss and deafness,” and “Leigh syndrome” (Figure 2, Figure 5, Figure 6, Table 4, Table 5). These results suggest that GEN may stimulate oxidative phosphorylation in the mitochondria and cause mitochondrial dysfunction, which is consistent with previous studies. In cytochrome c oxidase subunit 2 (COX2), G7598A mutation may aggravate mitochondrial dysfunction conferred by the A1555G mutation (Chen et al., 2013). Several cytochrome b (CYTB) mutations have been found in hearing-impaired Chinese pedigrees carrying the A1555G mutation (Yuan et al., 2005; Lu et al., 2010). Further in vitro and in vivo studies are necessary to elucidate the relationship between GEN-induced hearing loss and oxidative phosphorylation-associated genes identified using the DIAMOnD algorithm and the Cluster 1 genes found in this study.
4.2.2. Association between GEN and integrin genes
Integrin α8β1 and other integrins regulate hair cell differentiation and stereocilia maturation, and mutations in integrin genes may lead to inner ear diseases (Evans and Müller, 2000). Integrins both directly activate and enhance the growth factor activation of Rac and Cdc42 through membrane targeting. Rac and Cdc42 regulate activation of the JNK/stress-activated protein kinase (SAPK) and MAPK cascades (Coso et al., 1995; Minden et al., 1995; Schmidt and Hall, 1998). Kanamycin induces actin depolymerization, which is mediated by Rac1 activation, followed by superoxide formation by NADPH oxidase (Jiang et al., 2006). GEN-induced ototoxicity leads to JNK activation and apoptosis of inner hair cells (Ylikoski et al., 2002; Tao and Segil, 2015). After administering AGs, the sensory cells of the organ of Corti progressively degenerate, reducing the activity of mature brain-derived neurotrophic factor (BDNF) and inducing the activity of pro-BDNF, which leads to the upregulation of p75 neurotrophin receptor (NTR) and c-Jun thereby contributing to SGN death (Tan and Shepherd, 2006).
In the ClueGO analysis using KEGG, “PI3K-Akt signaling pathway,” “Ras signaling pathway,” “focal adhesion,” “MAPK signaling pathway,” “neurotrophin signaling pathway,” “regulation of actin cytoskeleton,” “apoptosis,” and “ECM-receptor interaction” were associated with GEN (Figure 3, Table 2, Table S2). Several GEN-associated genes based on DIAMOnD algorithm were highly enriched in “PI3K-Akt signaling pathway” (ATF2, ITGB1, ITGA3, EGFR, and HSP90B1), “Ras signaling pathway” (EGFR), “focal adhesion” (ITGB1, ITGA3, and EGFR), “MAPK signaling pathway” (ATF2 and EGFR), “regulation of actin cytoskeleton” (ITGB1, ITGA3, and EGFR), and “ECM-receptor interaction” (ITGB1 and ITGA3) (Figure 3, Figure 7, Table 2, Table 6, Table S2).
We found that several integrin genes (ITGA2, ITGA3, ITGB1, and ITGB4) may be associated with GEN (Figure 7, Table 6). We hypothesized that GEN may upregulate integrin genes, inducing actin depolymerization and MAPK cascades, which lead to hair cell apoptosis, and then upregulate p75NTR and SGN death. However, integrins contribute to the activation of PI3K, possibly via FAK and Cdc42, and promote polymerization and organization of actin filaments through both direct physical connections and a variety of signaling pathways (Schwartz and Assoian, 2001). Activation of PI3K signaling prevents AG-induced hair cell death (Jadali and Kwan, 2016). According to our findings, GEN might be associated with the PI3K-Act signaling pathway (Figure 3, Table 2, Table S2). However, the effect of GEN on the PI3K-Act signaling pathway cannot be explained by the upregulation of integrin genes. The downregulation of the PI3K-Act signaling pathway by GEN may be an integrin-independent pathway.
β1-integrin is a binding partner of EMILIN1 (Spessotto et al., 2003; Selvakumar et al., 2013), which in turn undergoes PPI with CNGA3 (Selvakumar et al., 2012). CNGA3 is a subunit of the cyclic nucleotide-gated (CNG) channel which represents a primary mechanism for Ca2+ entry and participates in hair cell mechanotransduction (Selvakumar et al., 2013). Mechanotransduction activity is important for the entry of AG into hair cells and is required for AG-induced ototoxicity (Marcotti et al., 2005; Alharazneh et al., 2011; Hailey et al., 2017). In this study, we found that some integrin genes (ITGA2, ITGA3, ITGB1, ITGB4) may be associated with GEN (Figure 7, Table 6). AG entry into hair cells may be associated with integrins.
4.3. Limitations
Our study has some limitations that should be considered. First, First, SRSs are passive reporting systems. The FAERS database is influenced by many factors such as over- or under-reporting, lack of a control population, missing data, and other confounders. Therefore, ROR can only indicate a possible causal relationship between AE and a drug and should not be applied to inferences regarding the comparative degrees of causality (van Puijenbroek et al., 2002). Second, drug–gene interactions were not demonstrated through in vitro and in vivo experiments in this study. In principle, this could be because of our currently limited knowledge of disease-associated proteins and their interactions. Therefore, the association between GEN and genes should be confirmed experimentally.
In the preliminary analysis of other AGs, we retrieved seed genes for kanamycin (MT-RNR1), tobramycin (MT-RNR1), amikacin (B2M, CSF2, IFNG, MT-RNR1, and SMN1), and neomycin (GYPA, MT-RNR1, PI3KCG, RLN2, and SLC2A4) from PharmGKB and DGIdb. The number of nodes and edges of the amikacin-associated gene network were 578 and 6835, respectively, while those of the neomycin-associated gene network were 519 and 33 386, respectively. In the ClueGO analysis using KEGG, “neurotrophin signaling pathway,” “PI3K-Akt signaling pathway,” and “Ras signaling pathway” were associated with amikacin. “Regulation of actin cytoskeleton” was associated with neomycin. Therefore, although the comparison of molecular-level functions, clinically relevant variants, and DIAMOnD analysis among AGs is interesting, we focused only on GEN in this study and did not investigate other AGs further.
Despite these limitations, we showed that several AGs may be associated with DIHL, which is consistent with previous research. We also showed that GEN may be associated with oxidative phosphorylation-associated genes and integrin genes, which may be associated with hearing loss. These data may provide insight into the toxicological mechanisms of GEN-induced hearing loss and further the development of drugs that prevent GEN-induced hearing loss.
5. Conclusion
Our analysis demonstrated an association between several AGs and DIHL using the FAERS. Drug–gene network analysis demonstrated that GEN may be associated with oxidative phosphorylation-associated genes and integrin genes, which may, in turn, be associated with hearing loss. Despite the limitations associated with SRS data and the lack of in vitro and in vivo experiments, we believe that our findings may provide insights into the toxicological mechanism of GEN-induced hearing loss and further the development of drugs that prevent GEN-induced hearing loss.
Declarations
Author contribution statement
Mizuki Tanaka; Mitsuhiro Nakamura: Conceived and designed the experiments; Performed the experiments; Analyzed and interpreted the data; Contributed reagents, materials, analysis tools or data; Wrote the paper.
Kiyoka Matsumoto; Riko Satake; Yu Yoshida; Misaki Inoue; Takaaki Suzuki; Mari Iwata: Performed the experiments; Analyzed and interpreted the data.
Shiori Hasegawa: Conceived and designed the experiments; Performed the experiments; Contributed reagents, materials, analysis tools or data.
Kazuhiro Iguchi: Conceived and designed the experiments; Wrote the paper.
Funding statement
This research was partially supported by the Japan Society for the Promotion of Science (KAKENHI grant number, 17K08452 and 20K10408).
Data availability statement
Data included in article/supplementary material/referenced in article.
Declaration of interests statement
The authors declare no conflict of interest.
Additional information
No additional information is available for this paper.
Appendix A. Supplementary data
The following is the supplementary data related to this article:
FigS1.
FigS2.
References
- Alharazneh A., Luk L., Huth M., Monfared A., Steyger P.S., Cheng A.G., Ricci A.J. Functional hair cell mechanotransducer channels are required for aminoglycoside ototoxicity. PloS One. 2011;6 doi: 10.1371/journal.pone.0022347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altissimi G., Colizza A., Cianfrone G., de Vincentiis M., Greco A., Taurone S., Musacchio A., Ciofalo A., Turchetta R., Angeletti D., Ralli M. Drugs inducing hearing loss, tinnitus, dizziness and vertigo: an updated guide. Eur. Rev. Med. Pharmacol. Sci. 2020;24:7946–7952. doi: 10.26355/eurrev_202008_22477. [DOI] [PubMed] [Google Scholar]
- Ashburner M., Ball C.A., Blake J.A., Botstein D., Butler H., Cherry J.M., Davis J.P., Dolinski K., Dwight S.S., Eppig J.T., Harris M.A., Hill D.P., Issel-Tarver L., Kasarskis A., Lewis S., Matese J.C., Richardson J.E., Ringwald M., Rubin G.M., Sherlock G., The Gene Ontology Consortium Gene ontology: tool for the unification of biology. Nat. Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bader G.D., Betel D., Hogue C.W.V. BIND: the biomolecular interaction network database. Nucleic Acids Res. 2003;31:248–250. doi: 10.1093/nar/gkg056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barabási A.L., Gulbahce N., Loscalzo J. Network medicine: a network-based approach to human disease. Nat. Rev. Genet. 2011;12:56–68. doi: 10.1038/nrg2918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bindea G., Mlecnik B., Hackl H., Charoentong P., Tosolini M., Kirilovsky A., Fridman W.H., Pagès F., Trajanoski Z., Galon J. ClueGO: a Cytoscape plug-in to decipher functionally grouped gene ontology and pathway annotation networks. Bioinformatics. 2009;25:1091–1093. doi: 10.1093/bioinformatics/btp101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyle E.A., Li Y.I., Pritchard J.K. An expanded view of complex traits: from polygenic to omnigenic. Cell. 2017;169:1177–1186. doi: 10.1016/j.cell.2017.05.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen T., Liu Q., Jiang L., Liu C., Ou Q. Mitochondrial COX2 G7598A mutation may have a modifying role in the phenotypic manifestation of aminoglycoside antibiotic-induced deafness associated with 12S rRNA A1555G mutation in a Han Chinese pedigree. Genet. Test. Mol. Biomarkers. 2013;17:122–130. doi: 10.1089/gtmb.2012.0251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coffin A.B., Rubel E.W., Raible D.W. Bax, Bcl2, and p53 differentially regulate neomycin- and gentamicin-induced hair cell death in the zebrafish lateral line. J. Assoc. Res. Otolaryngol. 2013;14:645–659. doi: 10.1007/s10162-013-0404-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coso O.A., Chiariello M., Yu J.C., Teramoto H., Crespo P., Xu N., Miki T., Gutkind J.S. The small GTP-binding proteins Rac1 and Cdc42 regulate the activity of the JNK/SAPK signaling pathway. Cell. 1995;81:1137–1146. doi: 10.1016/s0092-8674(05)80018-2. [DOI] [PubMed] [Google Scholar]
- Desa D.E., Nichols M.G., Smith H.J. Aminoglycosides rapidly inhibit NAD(P)H metabolism increasing reactive oxygen species and cochlear cell demise. J. Biomed. Opt. 2018;24:1–14. doi: 10.1117/1.JBO.24.5.051403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esterberg R., Linbo T., Pickett S.B., Wu P., Ou H.C., Rubel E.W., Raible D.W. Mitochondrial calcium uptake underlies ROS generation during aminoglycoside-induced hair cell death. J. Clin. Invest. 2016;126:3556–3566. doi: 10.1172/JCI84939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans A.L., Müller U. Stereocilia defects in the sensory hair cells of the inner ear in mice deficient in integrin α8β1. Nat. Genet. 2000;24:424–428. doi: 10.1038/74286. [DOI] [PubMed] [Google Scholar]
- Feissner R.F., Skalska J., Gaum W.E., Sheu S.S. Crosstalk signaling between mitochondrial Ca2+ and ROS. Front. Biosci. 2009;14:1197–1218. doi: 10.2741/3303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gatell J.M., Ferran F., Araujo V., Bonet M., Soriano E., Traserra J., SanMiguel J.G. Univariate and multivariate analyses of risk factors predisposing to auditory toxicity in patients receiving aminoglycosides. Antimicrob. Agents Chemother. 1987;31:1383–1387. doi: 10.1128/aac.31.9.1383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gene Ontology Consortium Gene ontology Consortium; going forward. Nucleic Acids Res. 2015;43:D1049–D1056. doi: 10.1093/nar/gku1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghiassian S.D., Menche J., Barabási A.L. A DIseAse MOdule Detection (DIAMOnD) algorithm derived from a systematic analysis of connectivity patterns of disease proteins in the human interactome. PLoS Comput. Biol. 2015;11 doi: 10.1371/journal.pcbi.1004120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goh K.I., Cusick M.E., Valle D., Childs B., Vidal M., Barabási A.L. The human disease network. Proc. Natl. Acad. Sci. U. S. A. 2007;104:8685–8690. doi: 10.1073/pnas.0701361104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goll J., Rajagopala S.V., Shiau S.C., Wu H., Lamb B.T., Uetz P. MPIDB: the microbial protein interaction database. Bioinformatics. 2008;24:1743–1744. doi: 10.1093/bioinformatics/btn285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Güldener U., Münsterkötter M., Oesterheld M., Pagel P., Ruepp A., Mewes H.W., Stümpflen V. MPact: the MIPS protein interaction resource on yeast. Nucleic Acids Res. 2006;34:D436–D441. doi: 10.1093/nar/gkj003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hailey D.W., Esterberg R., Linbo T.H., Rubel E.W., Raible D.W. Fluorescent aminoglycosides reveal intracellular trafficking routes in mechanosensory hair cells. J. Clin. Invest. 2017;127:472–486. doi: 10.1172/JCI85052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hobbie S.N., Akshay S., Kalapala S.K., Bruell C.M., Shcherbakov D., Böttger E.C. Genetic analysis of interactions with eukaryotic rRNA identify the mitoribosome as target in aminoglycoside ototoxicity. Proc. Natl. Acad. Sci. U. S. A. 2008;105:20888–20893. doi: 10.1073/pnas.0811258106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jadali A., Kwan K.Y. Activation of PI3K signaling prevents aminoglycoside-induced hair cell death in the murine cochlea. Biol. Open. 2016;5:698–708. doi: 10.1242/bio.016758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang H., Sha S.H., Schacht J. Rac/Rho pathway regulates actin depolymerization induced by aminoglycoside antibiotics. J. Neurosci. Res. 2006;83:1544–1551. doi: 10.1002/jnr.20833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang M., Karasawa T., Steyger P.S. Aminoglycoside-induced cochleotoxicity: a review. Front. Cell. Neurosci. 2017;11:308. doi: 10.3389/fncel.2017.00308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanehisa M., Goto S., Kawashima S., Nakaya A. The KEGG databases at GenomeNet. Nucleic Acids Res. 2002;30:42–46. doi: 10.1093/nar/30.1.42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanehisa M., Araki M., Goto S., Hattori M., Hirakawa M., Itoh M., Katayama T., Kawashima S., Okuda S., Tokimatsu T., Yamanishi Y. KEGG for linking genomes to life and the environment. Nucleic Acids Res. 2008;36:D480–D484. doi: 10.1093/nar/gkm882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landrum M.J., Lee J.M., Benson M., Brown G., Chao C., Chitipiralla S., Gu B., Hart J., Hoffman D., Hoover J., Jang W., Katz K., Ovetsky M., Riley G., Sethi A., Tully R., Villamarin-Salomon R., Rubinstein W., Maglott D.R. ClinVar: public archive of interpretations of clinically relevant variants. Nucleic Acids Res. 2016;44:D862–D868. doi: 10.1093/nar/gkv1222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanvers-Kaminsky C., Zehnhoff-Dinnesen A.A., Parfitt R., Ciarimboli G. Drug-induced ototoxicity: mechanisms, Pharmacogenetics, and protective strategies. Clin. Pharmacol. Ther. 2017;101:491–500. doi: 10.1002/cpt.603. [DOI] [PubMed] [Google Scholar]
- Lin Y., He S., Feng R., Xu Z., Chen W., Huang Z., Liu Y., Zhang Q., Zhang B., Wang K., Wu S. Digoxin-induced anemia among patients with atrial fibrillation and heart failure: clinical data analysis and drug-gene interaction network. Oncotarget. 2017;8:57003–57011. doi: 10.18632/oncotarget.18504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu J., Qian Y., Li Z., Yang A., Zhu Y., Li R., Yang L., Tang X., Chen B., Ding Y., Li Y., You J., Zheng J., Tao Z., Zhao F., Wang J., Sun D., Zhao J., Meng Y., Guan M.X. Mitochondrial haplotypes may modulate the phenotypic manifestation of the deafness-associated 12S rRNA 1555A>G mutation. Mitochondrion. 2010;10:69–81. doi: 10.1016/j.mito.2009.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ludovini V., Bianconi F., Siggillino A., Piobbico D., Vannucci J., Metro G., Chiari R., Bellezza G., Puma F., Della Fazia M.A., Servillo G., Crinò L. Gene identification for risk of relapse in stage I lung adenocarcinoma patients: a combined methodology of gene expression profiling and computational gene network analysis. Oncotarget. 2016;7:30561–30574. doi: 10.18632/oncotarget.8723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maiorino E., Baek S.H., Guo F., Zhou X., Kothari P.H., Silverman E.K., Barabási A.L., Weiss S.T., Raby B.A., Sharma A. Discovering the genes mediating the interactions between chronic respiratory diseases in the human interactome. Nat. Commun. 2020;11:811. doi: 10.1038/s41467-020-14600-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcotti W., van Netten S.M., Kros C.J. The aminoglycoside antibiotic dihydrostreptomycin rapidly enters mouse outer hair cells through the mechano-electrical transducer channels. J. Physiol. 2005;567:505–521. doi: 10.1113/jphysiol.2005.085951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menche J., Sharma A., Kitsak M., Ghiassian S.D., Vidal M., Loscalzo J., Barabási A.L. Uncovering disease-disease relationships through the incomplete interactome. Science. 2015;347:1257601. doi: 10.1126/science.1257601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minden A., Lin A., Claret F.X., Abo A., Karin M. Selective activation of the JNK signaling cascade and c-Jun transcriptional activity by the small GTPases Rac and Cdc42Hs. Cell. 1995;81:1147–1157. doi: 10.1016/s0092-8674(05)80019-4. [DOI] [PubMed] [Google Scholar]
- Pagel P., Kovac S., Oesterheld M., Brauner B., Dunger-Kaltenbach I., Frishman G., Montrone C., Mark P., Stümpflen V., Mewes H.W., Ruepp A., Frishman D. The MIPS mammalian protein-protein interaction database. Bioinformatics. 2005;21:832–834. doi: 10.1093/bioinformatics/bti115. [DOI] [PubMed] [Google Scholar]
- Prezant T.R., Agapian J.V., Bohlman M.C., Bu X., Öztas S., Qiu W.Q., Arnos K.S., Cortopassi G.A., Jaber L., Rotter J.I., Shohat M., Fischel-Ghodsian N. Mitochondrial ribosomal RNA mutation associated with both antibiotic–induced and non–syndromic deafness. Nat. Genet. 1993;4:289–294. doi: 10.1038/ng0793-289. [DOI] [PubMed] [Google Scholar]
- Raimundo N., Song L., Shutt T.E., McKay S.E., Cotney J., Guan M.X., Gilliland T.C., Hohuan D., Santos-Sacchi J., Shadel G.S. Mitochondrial stress engages E2F1 apoptotic signaling to cause deafness. Cell. 2012;148:716–726. doi: 10.1016/j.cell.2011.12.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Razick S., Magklaras G., Donaldson I.M. iRefIndex: a consolidated protein interaction database with provenance. BMC Bioinf. 2008;9:405. doi: 10.1186/1471-2105-9-405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt A., Hall M.N. Signaling to the actin cytoskeleton. Annu. Rev. Cell Dev. Biol. 1998;14:305–338. doi: 10.1146/annurev.cellbio.14.1.305. [DOI] [PubMed] [Google Scholar]
- Schwartz M.A., Assoian R.K. Integrins and cell proliferation: regulation of cyclin-dependent kinases via cytoplasmic signaling pathways. J. Cell Sci. 2001;114:2553–2560. doi: 10.1242/jcs.114.14.2553. [DOI] [PubMed] [Google Scholar]
- Selvakumar D., Drescher M.J., Dowdall J.R., Khan K.M., Hatfield J.S., Ramakrishnan N.A., Drescher D.G. CNGA3 is expressed in inner ear hair cells and binds to an intracellular C-terminus domain of EMILIN1. Biochem. J. 2012;443:463–476. doi: 10.1042/BJ20111255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selvakumar D., Drescher M.J., Drescher D.G. Cyclic nucleotide-gated channel α-3 (CNGA3) interacts with Stereocilia Tip-Link Cadherin 23 + Exon 68 or Alternatively with Myosin VIIa, two proteins required for hair cell mechanotransduction. J. Biol. Chem. 2013;288:7215–7229. doi: 10.1074/jbc.M112.443226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma A., Menche J., Huang C.C., Ort T., Zhou X., Kitsak M., Sahni N., Thibault D., Voung L., Guo F., Ghiassian S.D., Gulbahce N., Baribaud F., Tocker J., Dobrin R., Barnathan E., Liu H., Panettieri R.A., Jr., Tantisira K.G., Qiu W., Raby B.A., Silverman E.K., Vidal M., Weiss S.T., Barabási A.L. A disease module in the interactome explains disease heterogeneity, drug response and captures novel pathways and genes in asthma. Hum. Mol. Genet. 2015;24:3005–3020. doi: 10.1093/hmg/ddv001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma A., Kitsak M., Cho M.H., Ameli A., Zhou X., Jiang Z., Crapo J.D., Beaty T.H., Menche J., Bakke P.S., Santolini M., Silverman E.K. Integration of molecular interactome and targeted interaction analysis to identify a COPD disease network module. Sci. Rep. 2018;8:14439. doi: 10.1038/s41598-018-32173-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spessotto P., Cervi M., Mucignat M.T., Mungiguerra G., Sartoretto I., Doliana R., Colombatti A. β1 integrin-dependent cell adhesion to EMILIN-1 is mediated by the gC1q domain. J. Biol. Chem. 2003;278:6160–6167. doi: 10.1074/jbc.M208322200. [DOI] [PubMed] [Google Scholar]
- Su G., Morris J.H., Demchak B., Bader G.D. Biological network exploration with Cytoscape 3. Curr. Protoc. Bioinformatics. 2014;47:8–13. doi: 10.1002/0471250953.bi0813s47. 1–8.13.24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki Y., Suzuki H., Umetsu R., Uranishi H., Abe J., Nishibata Y., Sekiya Y., Miyamura N., Hara H., Tsuchiya T., Kinosada Y., Nakamura M. Analysis of the interaction between clopidogrel, aspirin, and proton pump inhibitors using the FDA Adverse Event Reporting System database. Biol. Pharm. Bull. 2015;38:680–686. doi: 10.1248/bpb.b14-00191. [DOI] [PubMed] [Google Scholar]
- Tan J., Shepherd R.K. Aminoglycoside-induced degeneration of adult spiral ganglion neurons involves differential modulation of tyrosine kinase B and p75 neurotrophin receptor signaling. Am. J. Pathol. 2006;169:528–543. doi: 10.2353/ajpath.2006.060122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka M., Hasegawa S., Nakao S., Shimada K., Mukai R., Matsumoto K., Nakamura M. Analysis of drug-induced hearing loss by using a spontaneous reporting system database. PloS One. 2019;14 doi: 10.1371/journal.pone.0217951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tao L., Segil N. Early transcriptional response to aminoglycoside antibiotic suggests alternate pathways leading to apoptosis in sensory hair cells in the mouse inner ear. Front. Cell. Neurosci. 2015;9:190. doi: 10.3389/fncel.2015.00190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Puijenbroek E.P., Bate A., Leufkens H.G.M., Lindquist M., Orre R., Egberts A.C.G. A comparison of measures of disproportionality for signal detection in spontaneous reporting systems for adverse drug reactions. Pharmacoepidemiol. Drug Saf. 2002;11:3–10. doi: 10.1002/pds.668. [DOI] [PubMed] [Google Scholar]
- Wang J., Van De Water T.R., Bonny C., de Ribaupierre F., Puel J.L., Zine A. A peptide inhibitor of c-Jun N-terminal kinase protects against both aminoglycoside and acoustic trauma-induced auditory hair cell death and hearing loss. J. Neurosci. 2003;23:8596–8607. doi: 10.1523/JNEUROSCI.23-24-08596.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wishart D.S., Knox C., Guo A.C., Shrivastava S., Hassanali M., Stothard P., Chang Z., Woolsey J. DrugBank: a comprehensive resource for in silico drug discovery and exploration. Nucleic Acids Res. 2006;34:D668–D672. doi: 10.1093/nar/gkj067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu M., Yu Q., Li Q. Differences in reproductive toxicology between alopecia drugs: an analysis on adverse events among female and male cases. Oncotarget. 2016;7:82074–82084. doi: 10.18632/oncotarget.12617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie J., Talaska A.E., Schacht J. New developments in aminoglycoside therapy and ototoxicity. Hear. Res. 2011;281:28–37. doi: 10.1016/j.heares.2011.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ylikoski J., Xing-Qun L., Virkkala J., Pirvola U. Blockade of c-Jun N-terminal kinase pathway attenuates gentamicin-induced cochlear and vestibular hair cell death. Hear. Res. 2002;166:33–43. doi: 10.1016/s0378-5955(01)00388-4. [DOI] [PubMed] [Google Scholar]
- Yu J., Zheng J., Zhao X., Liu J., Mao Z., Ling Y., Chen D., Chen C., Hui L., Cui L., Chen Y., Jiang P., Guan M.X. Aminoglycoside stress together with the 12S rRNA 1494C>T mutation leads to mitophagy. PloS One. 2014;9 doi: 10.1371/journal.pone.0114650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan H., Qian Y., Xu Y., Cao J., Bai L., Shen W., Ji F., Zhang X., Kang D., Mo J.Q., Greinwald J.H., Han D., Zhai S., Young W.Y., Guan M.X. Cosegregation of the G7444A mutation in the mitochondrial COI/tRNASer(UCN) genes with the 12S rRNA A1555G mutation in a Chinese family with aminoglycoside-induced and nonsyndromic hearing loss. Am. J. Med. Genet. A. 2005;138A:133–140. doi: 10.1002/ajmg.a.30952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao H., Li R., Wang Q., Yan Q., Deng J.H., Han D., Bai Y., Young W.Y., Guan M.X. Maternally inherited aminoglycoside-induced and nonsyndromic deafness is associated with the novel C1494T mutation in the mitochondrial 12S rRNA gene in a large Chinese family. Am. J. Hum. Genet. 2004;74:139–152. doi: 10.1086/381133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao S., Iyengar R. Systems pharmacology: network analysis to identify multiscale mechanisms of drug action. Annu. Rev. Pharmacol. Toxicol. 2012;52:505–521. doi: 10.1146/annurev-pharmtox-010611-134520. [DOI] [PMC free article] [PubMed] [Google Scholar]
Web references
- https://www.healthinaging.org/a-z-topic/hearing-loss
- https://www.who.int/news-room/fact-sheets/detail/deafness-and-hearing-loss
- http://www.fda.gov
- https://www.fda.gov/drugs/surveillance/questions-and-answers-fdas-adverse-event-reporting-system-faers
- https://www.drugbank.ca/
- https://www.pmrj.jp/jmo/php/indexj.php
- https://thebiogrid.org/
- http://mips.helmholtz-muenchen.de/corum/
- https://dip.doe-mbi.ucla.edu/dip/Main.cgi
- http://www.hprd.org/
- https://www.innatedb.com/
- https://www.ebi.ac.uk/intact/
- http://matrixdb.univ-lyon1.fr/
- https://mint.bio.uniroma2.it/
- https://reactome.org/
- http://virhostnet.prabi.fr/
- https://www.ebi.ac.uk/QuickGO/
- https://irefindex.vib.be/wiki/index.php/iRefIndex
- https://www.pharmgkb.org
- https://www.dgidb.org
- https://www.baderlab.org/Software/MCODE/UsersManual
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data included in article/supplementary material/referenced in article.