Abstract
Gut microbiota are critical to many aspects of human health including immune and metabolic health. Long-term diet influences the community structure and activity of the trillions of microorganisms residing in the human gut, but it remains unclear how the human gut microbiome responds to short-term intervention with dietary fiber. This study explored the effects of mixed dietary fibers on gut microbiota in young, healthy people. Twelve healthy, young adults participated in a randomized, crossover trial comparing the effects of polyglucan, inulin and resistant malt dextrin on gut microbiota composition and bacterial abundances. During the study, the subjects followed their normal diets without any constraints. Microbial community profiles were determined by absolute quantification 16S rRNA gene amplicon sequencing. Mixed model analysis did not reveal an effect of dietary intervention on microbial community structure. At the genus level, dietary fiber intervention for 4 days significantly promoted the growth of Alloprevotella, Parabacteroides and Parasutterella and inhibited the growth of Adlercreutzia, Anaerovorax, Enterococcus, Intestinibacter and Ruminococcus2 compared with the baseline. Addition of whey albumen powder for 4 days promoted the growth of Corynebacterium, Collinsella, Olsenella and Lactococcus but interfered with the growth of Megasphaera. Our results should be corroborated by randomized clinical trials with large sample size.
Keywords: dietary fiber, gut microbiota, short-term intervention
Introduction
Gut microbiota refers to the microorganisms harbored by the digestive tract of the human body and is regarded as one of the key organs of the human body.1 Our understanding of the biological roles of the gut microbiome, which include modulation of glucose and lipid metabolism,2 has increased dramatically over the past decade. Studies have shown that environmental factors such as diet and drugs have a greater impact on gut microbiota than genetic factors. The diversity of gut microbiota and the proportion of organisms belonging to certain genera are differ significantly according to the various dietary patterns of the host.3,4 Addition of prebiotics to the host diet can increase the abundance of microorganisms in certain families by more than 30%,5 and changes in gut microbiota can directly affect the metabolism of glucose and lipid;6 therefore, controlling the balance of the gut microbiota through dietary intervention may improve host metabolism and thereby reduce the onset of metabolic diseases. Adding dietary fiber to the diet is one of the most common dietary interventions for obese and diabetic patients.
Dietary fiber is defined as edible carbohydrate polymers with three or more monomeric units that are resistant to endogenous digestive enzymes. It is neither hydrolyzed nor absorbed in the small intestine.7 Dietary fiber, the seventh most important dietary nutrient, is associated with various diseases of human body, especially metabolic diseases.8 Multiple cohort studies have indicated that dietary fiber supplementation can improve the metabolism of subjects and reduce the risk of developing metabolic diseases such as obesity and type 2 diabetes.9–12
Current evidence regarding the effect of dietary fiber on the gut microbiota has been informed by specific prebiotic fiber interventions13, whole diet interventions14 and cross-sectional associations,15 yet there is no consistent conclusion on how dietary fiber alters the composition and diversity of gut microbiota since the microbial response to dietary fiber is highly individualized.16–19 More importantly, studies looking at the effects of dietary fiber on gut microbiota in healthy individuals, excluding infection or disease state, are limited. The influence of dietary fiber on the gut microbiota of healthy people may be even more important than that in unhealthy people, because intervention in healthy people before the occurrence of disease is the basis for preventing disease. Long-term dietary fiber intervention influences the structure and activity of the trillions of microorganisms residing in the human gut, but how rapidly the human gut microbiome responds to short-term dietary fiber intervention remains unclear. In this study, we conducted a randomized, controlled, cross-over trial involving a daily dietary fiber intervention in healthy volunteers. We evaluated the effect of mixed dietary fibers comprising polyglucan, inulin and resistant malt dextrin on gut microbiota composition and bacterial abundances among healthy volunteers.
Methods
Study Design and Participants
The study was registered as a clinical trial (http://www.chictr.org.cn/, China Clinical Trial Registration, ChiCTR1900027845) and carried out at Xinjiang Medical University (Urumqi, China). The study was conducted according to the Declaration of Helsinki. The Medical Ethics Committee of the First Affiliated Hospital of Xinjiang Medical University approved the study protocol (20191113-05). Participants were recruited through public advertisements. The protocol was explained to the volunteers, and their written informed consent was obtained. Inclusion criteria were as follows: age from 18 to 35; BMI [body weight (kg)/height (m2)] between 18 and 28; no intake of antibiotics, laxatives or other gastrointestinal medications for 3 months prior to the beginning of the study. A 4 mL venous blood sample was collected after at least 8 hours of fasting for blood glucose determination and blood and laboratory testing. The First Affiliated Hospital of Xinjiang Medical University performed blood glucose and blood lipid assays using auto-analyzers. The test methods and requirements complied with national clinical test operating procedures in China.
Twelve healthy, young, adults (six females and six males, aged from 22 to 32 years) with no history of gastrointestinal problems participated in the study. Subjects were randomly assigned to receive each dietary intervention during the 4-day treatment period with a 4-day washout period. Six subjects followed a first round of 20 g mixed dietary fibers per day for 4 days, followed by a 4-day washout, then a second round of 40 g whey albumen powder per day for 4 days. The other six participants followed the exact same sequence but with 40 g whey albumen powder per day in the first round and 20 g mixed dietary fibers per day in the second round. The mixed dietary fibers consisted of polyglucan, inulin and resistant malt dextrin (Revilife, China). Whey albumen powder was composed of hydrolyzed whey protein peptide (Revilife, China). Data from all subjects were separated into three groups: Group A represented data collected after every dietary fiber intervention. Group B represented data collected after every protein powder intervention. Group P represented summarized data from the washout period and baseline data.
During the study, the subjects followed their normal diet without any constraints. All diets were consumed ad libitum without caloric restrictions. Notepads were used to log diet and bowel movements during the experiment. The participants’ normal bowel frequencies ranged from three times a day to once every other day. All participants logged their diet for 3 days before the study began using the 24-hour diet review method. The Feihua nutrition calculator V2.7.5.4 (Feihua, China) was used to calculate the percentage of heat energy from carbohydrate, protein and fat. The subjects visited the Xinjiang Medical University five times to provide stool samples and were monitored by a clinical dietitian throughout the study. Metabolic measurements were made after overnight fasting.
Fecal Sample Collection and Storage
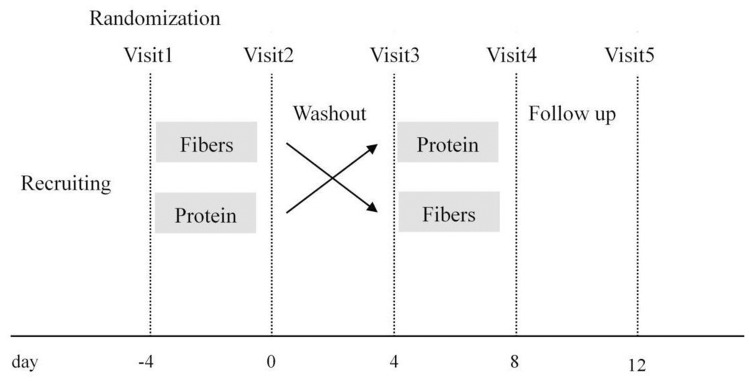
For each participant, five stool samples were taken during the study. The first sample was taken 4 days before the intervention start, the second sample was taken after the first-round intervention, the third sample was taken after the 4-day washout period, the fourth sample was taken after the second-round intervention, and the fifth sample was taken 4 days after the fourth sample. The stool sample collected for each period was from the first bowel movement during the stipulated time. The study design is shown in Figure 1. Some subjects were unable to defecate on the days of their visit, and a total of 52 stool samples were collected in this study. Stool samples for gut microbiota composition were collected in sterile tubes and placed into specimen containers; these were further surrounded by frozen gel packs cooled to −20°C, delivered to the laboratory in coolers within 2 h and then stored at −80°C.
Figure 1.
Intervention study design.
Microbial Community Analysis by 16S rRNA Gene Sequencing
Total DNA was extracted from all 52 stool samples using a QIAamp stool DNA mini kit (Qiagen, Germany) according to the manufacturer’s instructions. DNA was quantified using a NanoDrop ND-1000 spectrophotometer (Rockland Company, USA) and stored at −80°C for later use. The genomic DNA was used to construct an amplicon library by amplifying the V3~V4 region of the 16S rRNA gene. PCR was performed using the following conditions: initial denaturation at 95°C for 3 min; 25 cycles of 95°C for 30 s, 55°C for 30 s, and 72°C for 30 s; and a final extension step at 72°C for 7 min. After the reaction, all reaction products were detected using 1.5% agar gel electrophoresis (ethidium bromide staining) to determine the size of amplified fragments. A QIAquick GelExtraction Kit (QIAGEN, Germany) was used to recover and purify target bands. An Illumina Miseq high-throughput sequencing platform (Illumina) was used to sequence the PCR products in the 16S RNA V3~V4 region; the sequencing type was PE250. Reads with low quality were filtered out from the drop-out data to obtain clean data. The software FLASH (FastLength Adjustment of Short reads V1.2.11) was used for sequence stitching. Reads obtained from double-end sequencing were spliced into Tags. Optimised Tags were grouped into operational taxonomic units (OTUs) at 97% similarity, after obtaining the representative OTU sequence; representative OTU sequences were compared with known Greengene (V201305) sequences for species annotation through the Ribosomal Database Project Classifier (V2.2) software to obtain community composition information for each sample.
DNA of total bacteria in feces was extracted using a QIAamp® Fast DNA Stool Mini Kit. Absolute quantification by 16S rRNA gene pyrosequencing of the DNA samples was carried out by Shanghai Genesky Biotechnologies Inc. China. For each sample, the V3~V4 region of the bacterial 16S ribosomal RNA gene was chosen for amplification and sequenced on an Illumina MiSeq platform. Nine different spike-in sequences with four different concentrations (104, 105, 106, or 107 copies of internal standards) were added to the sample DNA pools. The spike-in sequences contained conserved regions identical to those of natural 16S rRNA genes and artificial variable regions different from nucleotide sequences in the public databases, which worked as internal standards and allowed absolute quantification across samples.20,21 All results were based on sequenced reads and operational taxonomic units (OTUs).
Statistical Analysis
Statistical analysis was carried out using SAS software (SAS 9.4; SAS Institute Inc. Cary, NC, USA). Results are presented as means ± SEMs, and log10 transformation was performed when necessary. Differences in absolute abundances of taxonomic groups were investigated at the levels of phylum, class, order, family, genus and species. The average absolute abundance of taxa in sample 1, sample 3 and sample 5 of the 12 participants were selected as baseline (group P). Comparisons were made between baseline and post-intervention for each diet intervention group and between the two diet intervention groups (group A and group B). Absolute abundances of taxa were analyzed using a mixed linear model with intervention group; the order of the interventions was used as the fixed effect, and subjects were used as the random effect. Differences in absolute taxon abundances between the two diet interventions and between the baseline and post-intervention, together with their 95% confidence intervals (CI), were derived from the mixed model. In all cases, the level of significance was set at 5%.
The α-diversity values for each sample were described with the use of the Chao1, Observed and Shannon diversity indices. The same method used to determine absolute taxon abundances was implemented for analyzing α-diversity. The β-diversity of intervention groups and baseline was also compared by performing principal co-ordinates analysis (PCoA) on weighted WuniFrac (a distance metric used for comparing biological communities) distances.
Results
Participant Characteristics
Twelve healthy, young adults (six males and six females) were involved in the current study; their baseline characteristics are provided in Table 1. None of them had a BMI beyond 28 kg/m2. The blood glucose, total cholesterol, triglyceride, high-density lipoprotein and low-density lipoprotein of all subjects were in the normal range.
Table 1.
Baseline Characteristics of Subjects
| Characteristic | Group: Fibers-Protein (Mean±SD; n=6) | Group: Protein-Fibers (Mean±SD; n=6) | Total (Mean±SD; n=12) |
|---|---|---|---|
| Age (year) | 27 (2.0) | 26 (4.0) | 27(3.0) |
| Sex (female/male) | 3/3 | 3/3 | 6/6 |
| Ethnicity | |||
| Han | 5 | 4 | 9 |
| Non-Han | 1 | 2 | 3 |
| Height (cm) | 166.3±5.6 | 170.3±6.3 | 168.3±6.1 |
| Weight (kg) | 61.4±14.1 | 64.1±13.3) | 62.8±13.2 |
| Waist circumference (cm) | 76.0±8.1 | 77.8±7.8 | 76.9±7.6 |
| Blood glucose (mmol/L) | 4.70±0.51 | 4.99±0.34 | 4.84±0.44 |
| Triglyceride (mmol/L) | 0.94±0.52 | 0.93±0.45 | 0.93±0.46 |
| Total cholesterol (mmol/L) | 4.04±0.56 | 4.52±0.34) | 4.27±0.68 |
| High density lipoprotein (mmol/L) | 1.38±0.36 | 1.46±0.34 | 1.42±0.34 |
| Low density lipoprotein (mmol/L) | 2.31±0.70 | 2.69±0.81 | 2.49±0.75 |
Food Logs and Dietary Questionnaires
Three repeated 24-hour dietary food lists were used to record the diet of all subjects before and after the study period. We analyzed the percentage of heat energy from carbohydrate, protein and fat among the 12 participants. The results are shown in Table 2.
Table 2.
Percentage of Heat Energy from Three Nutrients Among 12 Participants
| Characteristic | Group:Fibers-Protein | Group:Protein-Fibers | Total |
|---|---|---|---|
| Carbohydrate (percentage of energy) | 219.1(58.7%) | 197.1(59.2%) | 208.1(59.0%) |
| Protein (percentage of energy) | 64.0(18.2%) | 56.8(18.3%) | 60.4(18.2%) |
| Fat (percentage of energy) | 32.0(23.1%) | 41.4(22.5%) | 36.7(22.8%) |
Microbial Community Analysis
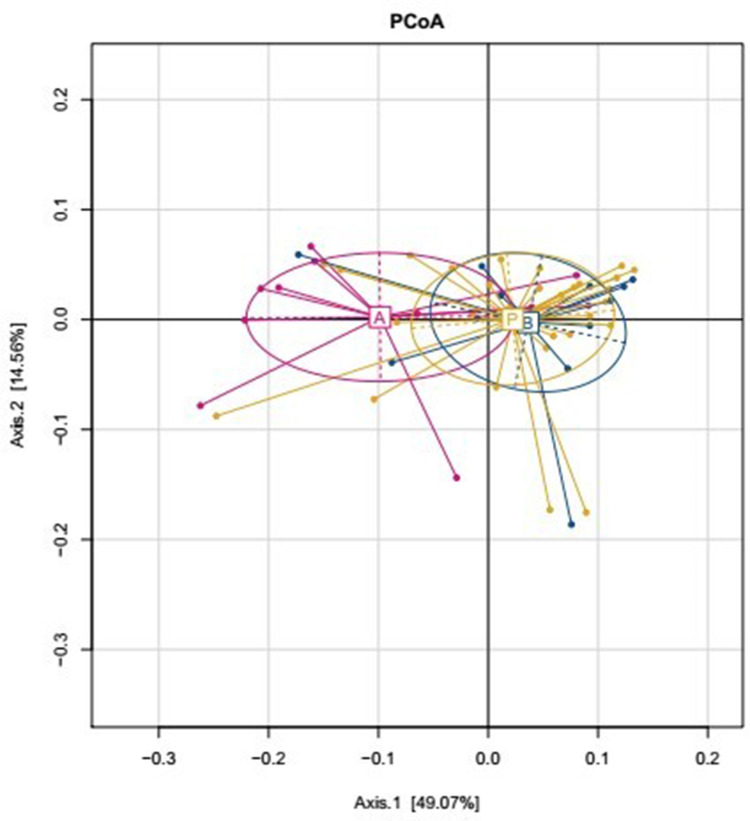
The effects of dietary fibers on the gut microbiota were determined by high-throughput sequencing analysis. We analyzed α-diversity indices [observed species and Chao 1 index (OTU richness estimation), and Shannon index (OTU evenness estimation)] using a mixed linear model. The results indicated only a slight difference in Observed, Chao1, and Shannon indices based on absolute quantification between the two diet interventions, and between baseline and post-intervention data (Table 3). We also compared the β-diversity between the two dietary interventions, and between the baseline and post-intervention samples by performing PCoA on weighted WuniFrac distances. The dietary fiber intervention group and the other groups were distinctly separate; however, there were only minor differences among the diet interventions, and among the baseline and post-intervention samples (Figure 2). In conclusion, mixed model analysis revealed that dietary interventions had no effects on microbial community structure.
Table 3.
Summary Statistics and Results from Mixed Model Analysis of α-Diversity Indices
| Index | Descriptive Statistics | Mixed Model Analysis | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Group | ||||||||||
| A (n=11)a | B (n=12)b | P (n=29)c | Total (n=52) | A vs P | B vs P | A vs B | ||||
| Difference (95% CI) | p value | Difference (95% CI) | p value | Difference (95% CI) | p value | |||||
| Observed | 378.73 (44.57) | 388.92 (50.82) | 389.52 (44.70) | 387.10 (45.42) | −8.45 (−30.98to14.08) | 0.452 | 2.36 (−19.36to24.08) | 0.828 | −10.81 (−37.23to15.62) | 0.413 |
| Chao1 | 461.73 (45.51) | 460.38 (59.22) | 471.61 (55.72) | 466.93 (53.82) | −8.16 (−39.30to22.97) | 0.599 | −7.95 (−37.99to22.09) | 0.595 | −0.21 (−36.76to36.33) | 0.990 |
| Shannon | 3.28 (0.36) | 3.51 (0.33) | 3.37 (0.34) | 3.38 (0.35) | −0.07 (−0.25to0.10) | 0.403 | 0.13 (−0.04to0.29) | 0.138 | −0.20 (−0.40to0.01) | 0.057 |
Notes: aGroup A represents data from all post-dietary fiber intervention treatments. bGroup B represents data from all post-protein powder intervention treatments. cGroup P represents summarized data for the washout periods and baselines.
Figure 2.
β-Diversity comparisons of gut microbiota in stool samples that were collected at baseline (group P) and after two dietary intervention groups (A: Dietary fibers group, B: Whey albumen powder group).
Bacterial composition was determined at the phylum, class, order, family, genus and species levels. At the genus level, compared with the baseline, 4 days of dietary fiber intervention boosted the growth of Alloprevotella, Parabacteroides and Parasutterella notably; the differences with 95% CI between dietary fiber intervention and baseline of log10 transformed absolute bacterial abundance were 0.70 (0.09 to 1.32), 2.68 (1.90 to 3.47) and 0.94 (0.14 to 1.74), with p values of 0.026, <0.001 and 0.022, respectively. Four days of dietary fiber intervention also dramatically prevented the growth of Adlercreutzia, Anaerovorax, Enterococcus, Intestinibacter and Ruminococcus2; the differences with 95% CI between dietary fiber intervention and baseline of log10 transformed absolute bacterial abundance were −1.14 (−1.72 to −0.55), −0.87 (−1.56 to −0.18), −1.78 (−3.22 to −0.33), −2.15 (−3.43 to −0.88) and −1.03 (−1.66 to −0.39), with p values of <0.001, 0.015, 0.018, 0.002 and 0.002, respectively. At the genus level, the change in log10 transformed absolute bacterial abundance between the 4 days of dietary fiber group and the 4 days of whey albumen powder group exhibited the same trend as the change in log10 transformed absolute bacterial abundance between the 4 days of dietary fiber group and the baseline. At the genus level, compared with the baseline, 4 days of whey albumen powder intervention fostered considerable growth of Corynebacterium, Collinsella, Olsenella and Lactococcus; the differences with 95% CI between 4 days of whey albumen powder intervention and baseline of log10 transformed absolute bacterial abundance were 1.36 (0.50 to 2.21),0.89 (0.33 to 1.44), 1.19 (0.25 to 2.12) and 2.67 (0.81 to 4.53), with p values of 0.003, 0.003, 0.014 and 0.006, respectively. At the genus level, compared with the baseline, 4 days of whey albumen powder intervention evidently impeded the growth of Megasphaera; the difference and 95% CI between 4 days of whey albumen intervention and baseline of log10 transformed absolute bacterial abundance was −1.07 (−2.03 to −0.11), p = 0.030 (Table 4).
Table 4.
Summary Statistics and Results from Mixed Model Analysis of Absolute Abundances of Genera
| Genus | Mean (SD) | Mixed Model Analysis | ||||
|---|---|---|---|---|---|---|
| A (n=11)a | B (n=12)b | P (n=29)c | Label | Difference (95% CI) | p value | |
| Adlercreutzia | 5.19(1.59) | 7.12(2.19) | 6.38(1.94) | A vs P | −1.14(−1.72to-0.55) | 0.000 |
| B vs P | 0.56(−0.01to1.12) | 0.053 | ||||
| A vs B | −1.70(−2.38to-1.01) | <0.001 | ||||
| Alloprevotella | 0.86(1.47) | 0.78(2.72) | 0.75(2.26) | A vs P | 0.70(0.09to1.32) | 0.026 |
| A vs B | 0.78(0.06to1.50) | 0.034 | ||||
| B vs P | −0.08(−0.67to0.51) | 0.792 | ||||
| Anaerovorax | 6.69(1.31) | 7.93(1.41) | 7.67(1.29) | A vs P | −0.87(−1.56to-0.18) | 0.015 |
| B vs P | 0.32(−0.34to0.99) | 0.336 | ||||
| A vs B | −1.19(−1.99to-0.38) | 0.005 | ||||
| Enterococcus | 2.75(2.47) | 4.63(2.45) | 4.39(2.35) | A vs P | −1.78(−3.22to-0.33) | 0.018 |
| B vs P | 0.11(−1.29to1.51) | 0.873 | ||||
| A vs B | −1.89(−3.59to-0.19) | 0.031 | ||||
| Intestinibacter | 4.67(1.39) | 7.50(1.85) | 6.81(1.98) | A vs P | −2.15(−3.43to-0.88) | 0.002 |
| B vs P | 0.72(−0.52to1.95) | 0.247 | ||||
| A vs B | −2.87(−4.37to-1.37) | 0.000 | ||||
| Parabacteroides | 12.68(1.53) | 9.83(1.37) | 10.10(1.18) | A vs P | 2.68(1.90to3.47) | <0.001 |
| A vs B | 2.92(2.00to3.85) | <0.001 | ||||
| B vs P | −0.24(−1.00to0.52) | 0.531 | ||||
| Parasutterella | 8.45(2.44) | 6.50(2.46) | 7.33(1.62) | A vs P | 0.94(0.14to1.74) | 0.022 |
| A vs B | 1.67(0.74to2.61) | 0.001 | ||||
| B vs P | −0.73(−1.50to0.04) | 0.061 | ||||
| Ruminococcus2 | 9.76(1.51) | 10.80(1.44) | 10.78(1.43) | A vs P | −1.03(−1.66to-0.39) | 0.002 |
| B vs P | 0.02(−0.59to0.63) | 0.956 | ||||
| A vs B | −1.04(−1.78to-0.30) | 0.007 | ||||
| Corynebacterium | 0.62(1.45) | 1.87(1.71) | 0.48(1.12) | A vs P | 0.11(−0.78to0.99) | 0.809 |
| B vs P | 1.36(0.50to2.21) | 0.003 | ||||
| A vs B | −1.25(−2.29to-0.21) | 0.020 | ||||
| Collinsella | 9.65(1.32) | 10.85(1.79) | 9.85(1.70) | A vs P | −0.23(−0.80to0.35) | 0.430 |
| B vs P | 0.89(0.33to1.44) | 0.003 | ||||
| A vs B | −1.11(−1.79to-0.44) | 0.002 | ||||
| Olsenella | 1.96(2.63) | 3.56(2.94) | 2.43(2.48) | A vs P | −0.56(−1.53to0.41) | 0.251 |
| B vs P | 1.19(0.25to2.12) | 0.014 | ||||
| A vs B | −1.74(−2.88to-0.61) | 0.004 | ||||
| Lactococcus | 2.40(2.25) | 5.91(2.37) | 3.22(2.94) | A vs P | −0.79(−2.71to1.12) | 0.407 |
| B vs P | 2.67(0.81to4.53) | 0.006 | ||||
| A vs B | −3.47(−5.73to-1.21) | 0.004 | ||||
| Megasphaera | 8.29(2.44) | 6.33(2.44) | 7.59(2.38) | A vs P | 0.64(−0.35to1.64) | 0.199 |
| B vs P | −1.07(−2.03to-0.11) | 0.030 | ||||
| A vs B | 1.72(0.55to2.89) | 0.005 | ||||
Notes: aGroup A represents data from all post-dietary fiber intervention treatments. bGroup B represents data from all post-protein powder intervention treatments. cGroup P represents summarized data for the washout periods and baselines.
Discussion
We observed the effect of dietary fiber on intestinal flora by adding dietary fiber to the diets of healthy individuals for 4 days, identified the bacterial species that showed notable changes in order to adapt to the intervention, and speculated on the cause of these changes.
At the taxonomic level, we found that after intervention, relative abundance of Parasutterella, Parabacteroides and Alloprevotella changed indicatively, with all bacteria showing an upward trend. In addition, the relative abundance of Enterococcus, Leuconostoc, Anaerovorax, Parvimonas Lachnospiraceae, Ruminococcus, Intestinibacter Adlercreutzia, Saccharibacteria and Raoultella were also affected by the intervention, showing a remarkable decline.
Numerous studies have shown that antibiotic use and dietary intervention (such as supplementation with probiotics and resistant starch) can cause changes in the structure of intestinal flora. Gut microbiota affects host health by producing short-chain fatty acids (SCFAs), altering amino acid metabolites and affecting bile acid metabolites.22,23
The genus of Parasutterella has been defined as a core component of human and mouse intestinal flora. It is one of the most frequently reported taxonomic units of intestinal betaproteobacteria, and in-depth studies over recent years have indicated that Parasutterella are associated with a variety of health outcomes.24–26 Ju27 used a colonization experiment of strain Parasutterella mc1 to prove that Parasutterella can affect the host physiology by altering metabolites. Succinate, one of the SCFAs produced by Parasutterella, can promote colonization of anaerobic bacteria, maintain homeostasis of gut microbiota and protect the host from infection.28 Succinate is also considered an agonist of FBPase,29 can promote gluconeogenesis and can improve glucose homeostasis and weight gain.30,31 Parasutterella also produces purine derivatives such as inosine, hypoxanthine and xanthine. These substances play a role in regulating host immunity and maintaining intestinal mucosal stability.32,33
Parabacteroides is recognized as part of the core intestinal flora34 and has important physiological functions in the host. Parabacteroides distasonis is the most studied strain of Parabacteroides in the human gut, with most studies showing that P. distasonis is relatively low in obese, non-alcoholic fatty liver and multiple sclerosis patients.35–37 Wang29 confirmed that P. distasonis has the ability to transform bile acid, produce succinic acid and promote the increase of bile acid in mice, regulating host metabolism. Other Parabacteroides isolated from the gut, such as Parabacteroides goldsteinii, have similar effects.
Alloprevotella is one of the normal gut microbes of the human body and is widely found in the oral cavity and digestive tract,38 maintaining stability. The relative abundance of Alloprevotella decreases obviously under conditions of disease. Appropriate administration, or prebiotic or probiotic supplementation, increases the abundance of Alloprevotella.39 In our study, the addition of dietary fiber significantly increased the abundance of Alloprevotella, which was consistent with existing research results. However, the specific process and mechanism of Alloprevotella function in the improvement of body health are still unknown.
Our intervention of dietary fiber not only increased the relative abundance of some bacteria, but also reduced the abundance of some bacteria, mainly those of phylum Firmicutes such as Enterococcus, Anaerovorax, Ruminococcus2 and Intestinibacter, and Adlercreutzia, which belongs to the phylum Actinobacteria. Firmicutes are the dominant flora in normal human intestine, accounting for about 50–75%, with Bacteroides representing 10–50% and Proteobacteria often less than 1%.40 Some changes in bacterial abundance are closely related to the health status of the host, for example, in obese hosts, the relative abundance of Firmicutes is higher than that in normal hosts, while the intestinal flora of healthy hosts shows higher numbers of Bacteroides.41,42 Increases in proportions of bacteria will change the stability of intestinal flora and lead to nutritional metabolism disorders, immune disorders and inflammatory response.25 Most of the bacteria that were reduced in our intervention were also susceptible in other studies; some of them are known to be harmful, but the influence of most bacteria with reduced abundance on host health status is still unclear.
It is well-acknowledged that the intestinal flora is an extremely complex ecosystem. Dietary fiber intervention alters the intestinal microbiota instead of only a single bacterial taxon. In our study, we found that dietary fiber intervention increased the relative abundance of beneficial bacteria significantly, including Parabacteroides, Parasutterella, which are related to glucose metabolism and lipid metabolism; meanwhile, the abundance of some harmful bacteria was reduced. Dietary fiber intervention regulated the structure of the gut microbiota. In the observation of group B, protein powder intervention practically counteracted the effects of dietary fiber intervention, but we are not clear what origin of these changes was and which groups of bacteria were changed by the direct action of intervention. Human intestinal flora exists with mutual restriction and interdependence according to community structure and proportion with the host. In further work, we will explore changes in the metabolic functions of the intestinal flora during intervention to enhance our understanding of the impact of dietary intervention on gut microbiota and host health.
Strengths and Limitations
The current evidence regarding the effect of dietary fiber on the gut microbiota is informed from specific prebiotic fiber interventions, whole diet interventions and cross-sectional associations, but there still was not a consistent conclusion on how dietary fiber alters the composition and diversity of gut microbiota. In this study, we conducted a randomized, controlled, cross-over trial with a daily dietary fiber intervention in healthy volunteers. The aim of the study was to evaluate the effect of mixed dietary fibers consisting of polyglucan, inulin and resistant malt dextrin on gut microbiota composition and bacterial abundances among healthy volunteers. We assessed the effect of dietary fiber on intestinal flora by adding dietary fiber to the diets of healthy individuals for 4 days and identified the bacterial species showing significant changes in order to adapt to the intervention.
We acknowledge that our study had some limitations. First, the sample size of our study was only 12 healthy adults, which was relatively small. In the future, sample size should be further expanded to obtain more stable results. Second, significant changes were observed after the short-term intervention. Intestinal flora changed after the intervention, but no further follow-up was conducted to observe the duration of the influence of dietary fiber and protein powder on intestinal flora. Third, we did not justify if a 4 days washout period is enough within this study design. Furthermore, baseline fiber intake is not reported from the individuals. Beneficial effects of dietary fiber on intestinal flora were observed in this study, but we did not explore the potential benefit of this change in the intestinal flora; future studies should further explore the mechanism of how dietary fiber affects intestinal flora. Due to these limitations, the findings should be interpreted with caution.
Conclusion
Dietary fiber intervention significantly promoted the growth of Alloprevotella, Parabacteroides and Parasutterella, and inhibited the growth of Adlercreutzia, Anaerovorax, Enterococcus, Intestinibacter and Ruminococcus2 at the genus level. The findings should be interpreted with caution due to the limited sample size, our results should be corroborated by randomized clinical trials with large sample size.
Acknowledgments
We thank our volunteers for their great effort in participating in this clinical trial.
Funding Statement
This research was supported by the “State Key Laboratory of Pathogenesis, Prevention and Treatment of Central Asian High Incidence Diseases” fund (no. SKL-HIDCA-2018-18) and Xinjiang Uygur Autonomous Region “13th Five-Year” Key Discipline (Plateau discipline)-Public Health and Preventive Medicine.
Data Sharing Statement
The study data are not freely available, but specific proposals for future collaboration are welcome, addressed to corresponding author (Dai Jianghong, Email: epdjh@163.com).
Author Contributions
All authors made a significant contribution to the work reported, whether that is in the conception, study design, execution, acquisition of data, analysis and interpretation, or in all these areas; took part in drafting, revising or critically reviewing the article; gave final approval of the version to be published; have agreed on the journal to which the article has been submitted; and agree to be accountable for all aspects of the work.
Disclosure
None of the authors has any conflicts of interest to declare.
References
- 1.Bull MJ, Plummer NT. Part 1: the human gut microbiome in health and disease. Integr Med. 2014;13(6):17–22. [PMC free article] [PubMed] [Google Scholar]
- 2.Wang Y, Kuang Z. The intestinal microbiota regulates body composition through NFIL3 and the circadian clock. Science. 2017;357(6354):912–916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Singh RK, Chang HW, Yan D, et al. Influence of diet on the gut microbiome and implications for human health. J Transl Med. 2017;15(1):73. doi: 10.1186/s12967-017-1175-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rothschild D, Weissbrod O, Barkan E, et al. Environment dominates over host genetics in shaping human gut microbiota. Nature. 2018;555(7695):210–215. doi: 10.1038/nature25973 [DOI] [PubMed] [Google Scholar]
- 5.Liu X, Cao S, Zhang X. Modulation of gut microbiota-brain axis by probiotics, prebiotics, and diet. J Agric Food Chem. 2015;63(36):7885–7895. doi: 10.1021/acs.jafc.5b02404 [DOI] [PubMed] [Google Scholar]
- 6.Dinan TG, Cryan JF. The microbiome-gut-brain axis in health and disease. Gastroenterol Clin North Am. 2017;46(1):77–89. doi: 10.1016/j.gtc.2016.09.007 [DOI] [PubMed] [Google Scholar]
- 7.DeVries JW. On defining dietary fibre. Proc Nutr Soc. 2003;62(1):37–43. doi: 10.1079/PNS2002234 [DOI] [PubMed] [Google Scholar]
- 8.Weickert MO, Pfeiffer AF. Metabolic effects of dietary fiber consumption and prevention of diabetes. J Nutr. 2008;138(3):439–442. doi: 10.1093/jn/138.3.439 [DOI] [PubMed] [Google Scholar]
- 9.Joanne S. Fiber and prebiotics: mechanisms and health benefits. Nutrients. 2013;5(4):1417–1435. doi: 10.3390/nu5041417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang Y, Duan Y, Zhu L, et al. Whole grain and cereal fiber intake and the risk of type 2 diabetes: a meta-analysis. Int J Mol Epidemiol Genet. 2019;10(3):38–46. [PMC free article] [PubMed] [Google Scholar]
- 11.Evans CEL. Dietary fibre and cardiovascular health: a review of current evidence and policy. Proc Nutr Soc. 2019;79(1):61–67. [DOI] [PubMed] [Google Scholar]
- 12.Rakvaag E, Fuglsang-Nielsen R, Bach Knudsen KE, et al. Whey protein combined with low dietary fiber improves lipid profile in subjects with abdominal obesity: a randomized, controlled trial. Nutrients. 2019;11(9):2091. doi: 10.3390/nu11092091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Roberfroid M, Gibson GR, Hoyles L, et al. Prebiotic effects: metabolic and health benefits. Br J Nutr. 2010;104(Suppl 2):S1–S63. doi: 10.1017/S0007114510003363 [DOI] [PubMed] [Google Scholar]
- 14.David LA, Maurice CF, Carmody RN, et al. Diet rapidly and reproducibly alters the human gut microbiome. Nature. 2014;505(7484):559–563. doi: 10.1038/nature12820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schnorr SL, Candela M, Rampelli S, et al. Gut microbiome of the hadza hunter-gatherers. Nat Commun. 2014;5(1):3654. doi: 10.1038/ncomms4654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Davis LMG, Martinez I, Walter J, Goin C, Hutkins RW, Tan P. Barcoded pyrosequencing reveals that consumption of galactooligosaccharides results in a highly specific bifidogenic response in humans. PLoS One. 2011;6(9):e25200. doi: 10.1371/journal.pone.0025200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Martinez I, Kim J, Duffy PR, Schlegel VL, Walter J, Heimesaat MM. Resistant starches types 2 and 4 have differential effects on the composition of the fecal microbiota in human subjects. PLoS One. 2010;5(11):e15046. doi: 10.1371/journal.pone.0015046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Walker AW, Ince J, Duncan SH, et al. Dominant and diet-responsive groups of bacteria within the human colonic microbiota. ISME J. 2011;5(2):220–230. doi: 10.1038/ismej.2010.118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.An R, Wilms E, Smolinska A, et al. Sugar beet pectin supplementation did not alter profiles of fecal microbiota and exhaled breath in healthy young adults and healthy elderly. Nutrients. 2019;11(9). doi: 10.3390/nu11092193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jiang SQ, Yu YN, Gao RW, et al. High-throughput absolute quantification sequencing reveals the effect of different fertilizer applications on bacterial community in a tomato cultivated coastal saline soil. Sci Total Environ. 2019;687:601–609. doi: 10.1016/j.scitotenv.2019.06.105 [DOI] [PubMed] [Google Scholar]
- 21.Mou J, Li Q, Shi W, Qi X, Song W, Yang J. Chain conformation, physicochemical properties of fucosylated chondroitin sulfate from sea cucumber Stichopus chloronotus and its in vitro fermentation by human gut microbiota. Carbohydr Polym. 2020;228:115359. doi: 10.1016/j.carbpol.2019.115359 [DOI] [PubMed] [Google Scholar]
- 22.Maslowski KM, Vieira AT, Ng A, et al. Regulation of inflammatory responses by gut microbiota and chemoattractant receptor GPR43. Nature. 2009;461(7268):1282–1286. doi: 10.1038/nature08530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Song X, Sun X, Oh SF, et al. Microbial bile acid metabolites modulate gut RORγ+ regulatory T cell homeostasis. Nature. 2020;577(7790):410–415. doi: 10.1038/s41586-019-1865-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Morotomi M. The family sutterellaceae. In: Rosenberg EDE, Lory S, Stackebrandt E, Thompson F, editors. The Prokaryotes. Berlin, Heidelberg: Springer; 2014:1005–1012. [Google Scholar]
- 25.Shin NR, Whon TW, Bae JW. Proteobacteria: microbial signature of dysbiosis in gut microbiota. Trends Biotechnol. 2015;33(9):496–503. doi: 10.1016/j.tibtech.2015.06.011 [DOI] [PubMed] [Google Scholar]
- 26.Chen YJ, Wu H, Wu SD, et al. Parasutterella, in association with irritable bowel syndrome and intestinal chronic inflammation. J Gastroenterol Hepatol. 2018;33(11):1844–1852. doi: 10.1111/jgh.14281 [DOI] [PubMed] [Google Scholar]
- 27.Ju T, Kong JY, Stothard P, Willing BP. Defining the role of parasutterella, a previously uncharacterized member of the core gut microbiota. ISME J. 2019;13(6):1520–1534. doi: 10.1038/s41396-019-0364-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kim YG, Sakamoto K, Seo SU, et al. Neonatal acquisition of clostridia species protects against colonization by bacterial pathogens. Science. 2017;356(6335):315–319. doi: 10.1126/science.aag2029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang K, Liao M, Zhou N, et al. Parabacteroides distasonis alleviates obesity and metabolic dysfunctions via production of succinate and secondary bile acids. Cell Rep. 2019;26(1):222–235 e225. doi: 10.1016/j.celrep.2018.12.028 [DOI] [PubMed] [Google Scholar]
- 30.De Vadder F, Kovatcheva-Datchary P, Zitoun C, Duchampt A, Backhed F, Mithieux G. Microbiota-produced succinate improves glucose homeostasis via intestinal gluconeogenesis. Cell Metab. 2016;24(1):151–157. doi: 10.1016/j.cmet.2016.06.013 [DOI] [PubMed] [Google Scholar]
- 31.de Vadder F, Mithieux G. Gut-brain signaling in energy homeostasis: the unexpected role of microbiota-derived succinate. J Endocrinol. 2018;236(2):R105–R108. doi: 10.1530/JOE-17-0542 [DOI] [PubMed] [Google Scholar]
- 32.He B, Hoang TK, Wang T, et al. Resetting microbiota by Lactobacillus reuteri inhibits T reg deficiency-induced autoimmunity via adenosine A2A receptors. J Exp Med. 2017;214(1):107–123. doi: 10.1084/jem.20160961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cui HX, Hu YN, Li JW, Yuan K. Hypoglycemic mechanism of the berberine organic acid salt under the synergistic effect of intestinal flora and oxidative stress. Oxid Med Cell Longev. 2018;2018:8930374. doi: 10.1155/2018/8930374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Falony G, Joossens M, Vieira-Silva S, et al. Population-level analysis of gut microbiome variation. Science. 2016;352(6285):560–564. doi: 10.1126/science.aad3503 [DOI] [PubMed] [Google Scholar]
- 35.Cekanaviciute E, Yoo BB, Runia TF, et al. Gut bacteria from multiple sclerosis patients modulate human T cells and exacerbate symptoms in mouse models. Proc Natl Acad Sci U S A. 2017;114(40):10713–10718. doi: 10.1073/pnas.1711235114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Del Chierico F, Nobili V, Vernocchi P, et al. Gut microbiota profiling of pediatric nonalcoholic fatty liver disease and obese patients unveiled by an integrated meta-omics-based approach. Hepatology. 2017;65(2):451–464. doi: 10.1002/hep.28572 [DOI] [PubMed] [Google Scholar]
- 37.Verdam FJ, Fuentes S, de Jonge C, et al. Human intestinal microbiota composition is associated with local and systemic inflammation in obesity. Obesity. 2013;21(12):E607–E615. doi: 10.1002/oby.20466 [DOI] [PubMed] [Google Scholar]
- 38.Chen J, Yu B, Chen D, et al. Changes of porcine gut microbiota in response to dietary chlorogenic acid supplementation. Appl Microbiol Biotechnol. 2019;103(19):8157–8168. doi: 10.1007/s00253-019-10025-8 [DOI] [PubMed] [Google Scholar]
- 39.Ding S, Ma Y, Liu G, Yan W, Jiang H, Fang J. Lactobacillus brevis alleviates DSS-induced colitis by reprograming intestinal microbiota and influencing serum metabolome in murine model. Front Physiol. 2019;10:1152. doi: 10.3389/fphys.2019.01152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Manichanh C, Borruel N, Casellas F, Guarner F. The gut microbiota in IBD. Nat Rev Gastroenterol Hepatol. 2012;9(10):599–608. doi: 10.1038/nrgastro.2012.152 [DOI] [PubMed] [Google Scholar]
- 41.Backhed F, Ding H, Wang T, et al. The gut microbiota as an environmental factor that regulates fat storage. Proc Natl Acad Sci U S A. 2004;101(44):15718–15723. doi: 10.1073/pnas.0407076101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ley RE, Turnbaugh PJ, Klein S, Gordon JI. Microbial ecology - human gut microbes associated with obesity. Nature. 2006;444(7122):1022–1023. doi: 10.1038/4441022a [DOI] [PubMed] [Google Scholar]