Fig. 6. NF-κB and Notch Can be Therapeutically Targeted in ALT-driven Cancer Cells.
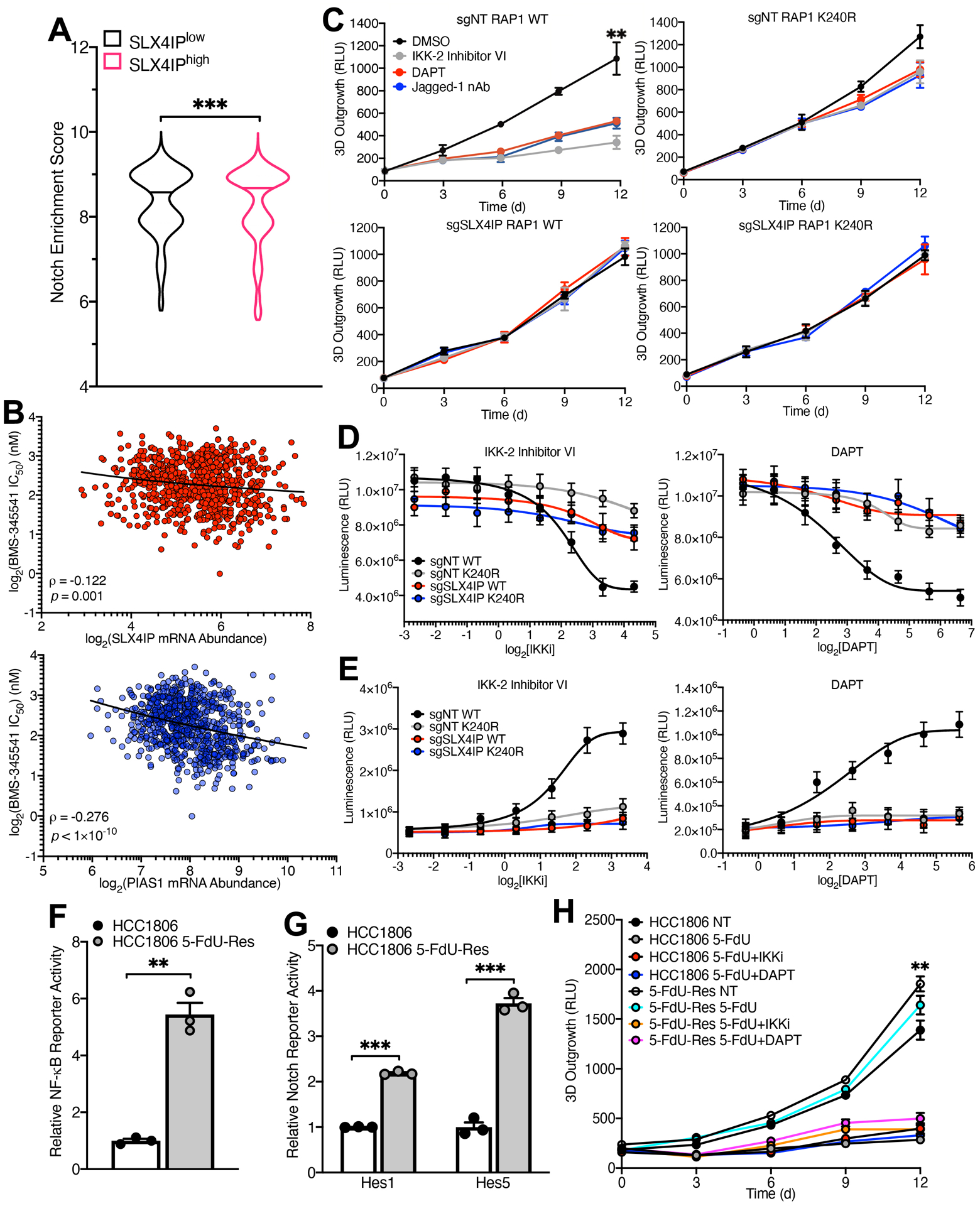
(A) Violin plot depicting pan-cancer enrichment analysis of a Notch-associated gene signature in TCGA specimens expressing high (SLX4IPhigh) and low (SLX4IPlow) levels of SLX4IP. Horizontal lines represent the median enrichment score for each group. n=4259 (SLX4IPlow), n=4207 (SLX4IPhigh). (B) Correlation (represented by Spearman’s ρ) of IKK inhibitor (BMS-345541) sensitivity with SLX4IP (top) and PIAS1 (bottom) expression across cancer cell lines (n=727). A lower IC50 value indicates increased sensitivity to treatment with BMS-345541. (C) Three-dimensional outgrowth of parental (top row) and SLX4IP-depleted (bottom row) U2OS cells expressing wild-type (left column) or SUMO-deficient (right column) RAP1. Cells were treated with diluent (DMSO), IKK-2 Inhibitor VI (1 μM), DAPT (25 μM), or Jagged-1 neutralizing antibody (nAb; 20 ng/mL) as indicated. n=3 biological replicates. (D) Viability of indicated U2OS derivatives treated with IKK-2 Inhibitor VI (left) or DAPT (right). n=3 independent experiments. (E) Caspase-3/7 activation in indicated U2OS derivatives treated as indicated. n=3 independent experiments. (F and G) NF-κB (F) and Notch (G) reporter activity in parental and 5-FdU-resistant (5-FdU-Res) HCC1806 cells. n=3 biological replicates for each. (H) Three-dimensional outgrowth of parental and 5-FdU-resistant HCC1806 cells treated with 5-FdU (500 nM) in combination with IKK-2 Inhibitor VI (1 μM) or DAPT (20 μM). n=3 biological replicates. **P<0.01 and ***P<0.001 by Mann-Whitney U test (A), Welch’s t test (F and G), or one-way ANOVA (C and H).
