FIGURE 4.
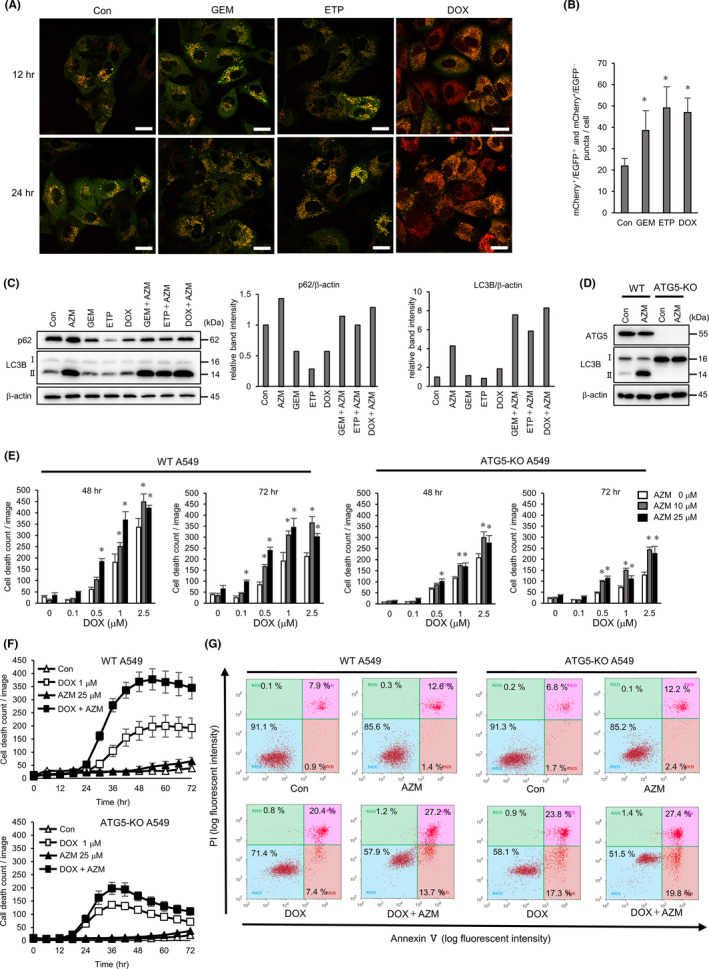
Azithromycin (AZM) enhanced cell toxicity by suppressing DNA‐damaging drug‐induced autophagy. A, Confocal microscopy of A549 cells expressing mCherry‐EGFP‐LC3 after treatment with gemcitabine (GEM) (1 µM), etoposide (ETP) (5 µM), or doxorubicin (DOX) (1 µM) for 12 or 24 h. Representative images of each treatment are shown. Scale bar = 20 µm. B, Number of red puncta (mCherry+ /EGFP‐ and mCherry+ /EGFP+) in each treated cells at 24 h is summarized. n = 5 (fields), bar = mean ± SD, *P < .05. vs control treatment. C, Western blotting of p62 and LC3B protein expression in A549 cells treated with indicated drugs to evaluate autophagy flux. Band intensity of p62 and LC3B‐II was measured and standardized by β‐actin expression and summarized in right columns. D, Western blotting analysis of ATG5‐KO in A549 cells. Expression of ATG5‐ATG12 and LC3B were analyzed after the control or 25 µM AZM treatment for 24 h. E, Wild‐type (WT) and ATG5‐KO A549 cells were treated with DOX ± AZM for up to 72 h. Number of dead cells was assessed by IncuCyte with propidium iodide (PI) staining. Representative data of three independent experiments are shown. n = 4, bar = mean ± SD, *P < .05 vs 0 µM AZM treatment. F, Time dependency of cell death shown in (E) is summarized. G, Flow cytometry analysis with the Annexin V/PI double‐stained WT or ATG5‐KO A549 cells after DOX (1 µM) ± AZM (25 µM) treatment for 72 h. Numbers indicate the percentage of the cell numbers in each area. Representative data of three independent experiments are shown
