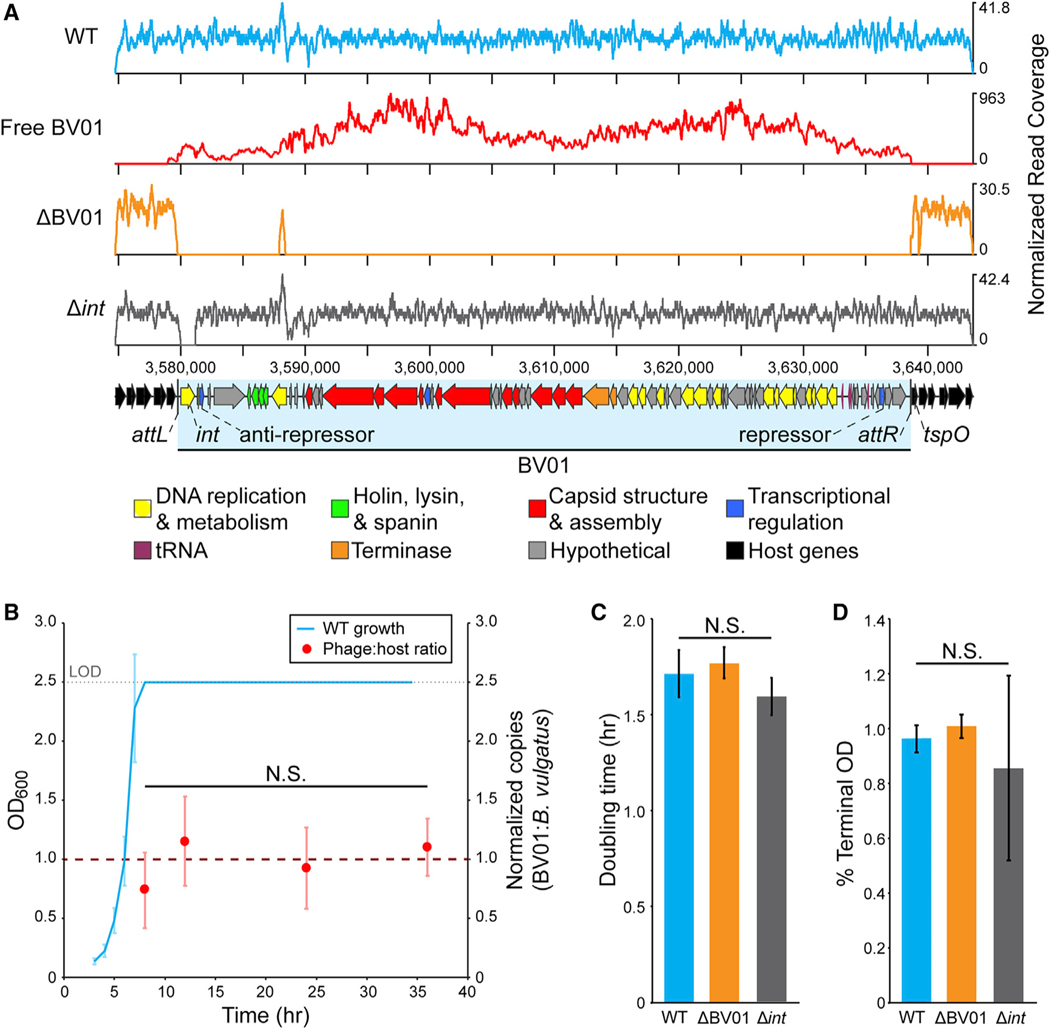
Figure 1. Phage BV01 Is Spontaneously Induced in Culture.
(A) DNA sequencing reads mapped to the BV01 prophage region. Reads from sequencing of B. vulgatus genomic DNA (WT) or isolated phage DNA (free BV01) were normalized to the total number of reads after trimming and are represented as a coverage curve. A cured lysogen (ΔBV01) and integrase deletion mutant (Δint) of B. vulgatus were confirmed by shotgun sequencing of genomic DNA (Tables S2 and S3). The coverage peak at position ~3,588,000 nt in ΔBV01 is attributed to a homologous sequence elsewhere in the B. vulgatus genome. Putative functional categories of BV01 genes are indicated by color; see Table S1 for full BV01 gene annotation.
(B) BV01 is produced at low levels at all stages of host growth. WT cultures were grown in triplicate for 36 h, and BV01 and host abundance were monitored by qPCR. By log phase, cultures reached the limit of detection (LOD) of the spectrophotometer.
(C and D) Lysogeny with phage BV01 does not affect (C) doubling time or (D) terminal optical density (OD) in vitro. Doubling times and terminal ODs were calculated for wild-type (WT), cured lysogen (ΔBV01), and integrase deletion (Δint) strains and averaged across three replicates.
Statistical analyses were performed with one-way ANOVA. N.S., not significant. Phage:host ratio df = 3, F = 4.03, p = 0.051. Doubling time df = 2, F = 2.55, p = 0.157. Percent terminal OD df = 2, F = 6.59, p = 0.101. All error bars represent standard deviation.