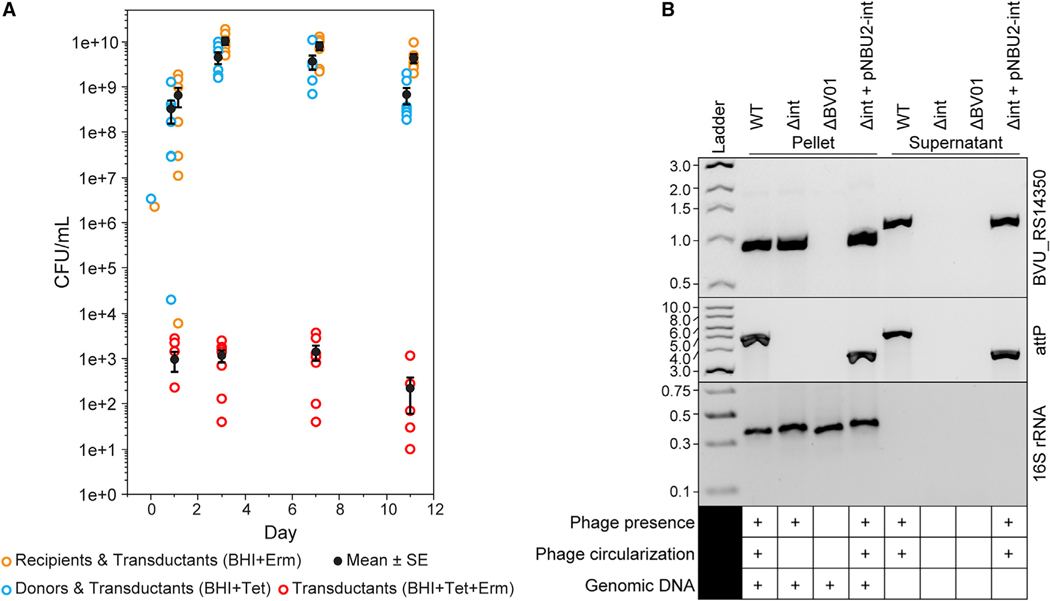
Figure 2. Transduction, Excision, and Circularization of Phage BV01.
(A) BV01 can transduce uninfected hosts in gnotobiotic mice (n = 7). Mice were gavaged with an equal mixture of BV01-tetQ lysogen and erythromycin-tagged cured lysogen (day 0). Recipient, donor, and transductant cells were identified by plating on medium with erythromycin (Erm) or tetracycline (Tet). Calculated means and standard errors (SE) are shown in black.
(B) Excision and circularization activities of the BV01 integrase are confirmed by PCR from late stationary phase cultures of four strains: WT, Δint, ΔBV01, and the int complement strain using a pNBU2 integrative plasmid (Δint + pNBU2-int). The presence of phage DNA was detected by amplification of a phage marker gene (BVU_RS14350). Amplification across the phage attachment site (attP) indicates circularization of the BV01 genome. Note that attP amplicons from Δint + pNBU2-int are ~1.2 kb shorter than WT amplicons because of deletion of the integrase gene. Supernatant fractions were DNase treated, eliminating all contaminating host genomic DNA, as demonstrated by lack of amplification of a host marker gene (16S rRNA). Despite an apparent size shift of BVU_RS14350 amplicons from the pellets and supernatants, Sanger sequencing validated that the products have no apparent insertions (Figure S1B). PCR amplicons were visualized by agarose gel electrophoresis alongside the New England Biolabs (NEB) 1-kb DNA ladder (BVU_RS14350, attP) or GeneRuler Express DNA ladder (16S rRNA); numbers are in kilobases.