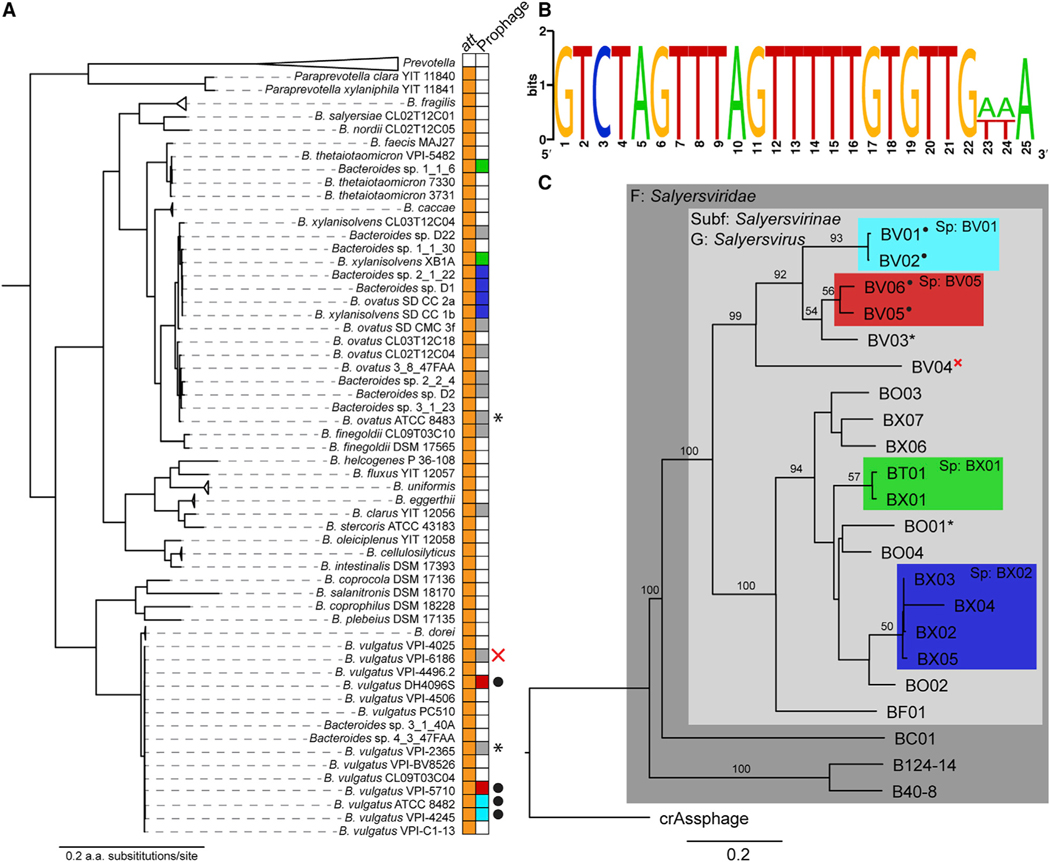
Figure 6. Salyersviridae Occur throughout the Bacteroides Genus.
(A) Bacteroides phylogeny and occurrence of the Salyersviridae att site. All duplications of the att site are associated with a putative integrated prophage. Host phylogeny was estimated by maximum likelihood from concatenated alignment of 13 core genes.
(B) Consensus att site for Salyersvirinae. The attP is duplicated upon integration of a Salyersvirinae prophage, resulting in direct repeats. The image was made with the WebLogo online tool.
(C) Phylogenomic genome-BLAST distance phylogeny implemented with the VICTOR online tool (Meier-Kolthoff and Göker, 2017) using amino acid data from all phage ORFs. For consistency, all phage genomes were annotated with MetaGeneAnnotator (Noguchi et al., 2008) implemented via VirSorter (Roux et al., 2015). Support values above branches are Genome Blast Distance Phylogeny (GBDP) pseudo-bootstrap values from 100 replications. Family (F), subfamily (Subf), genus (G), and species (Sp) were assigned by OPTSIL clustering (Gker et al., 2009; Table 1). Each leaf of the tree represents a unique phage species, except where indicated by colored boxes. Active prophages were confirmed by sequencing and/or PCR, indicated by black dots; prophages confirmed to have been inactivated by genome rearrangement are indicated by an X; prophages that were tested for activity with inconclusive results are indicated by asterisks (Figure S4).