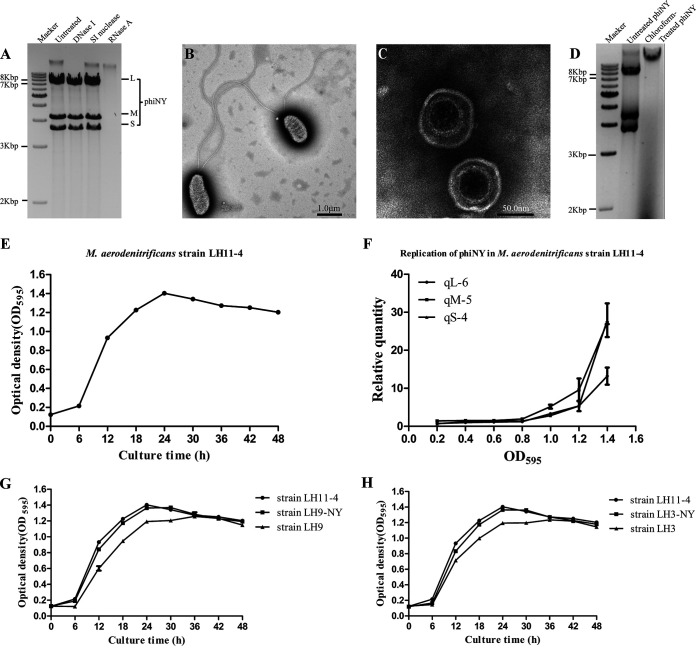
FIG 1.
Biological characterization of bacteriophage phiNY and the host bacterium Microvirgula aerodenitrificans strain LH11-4. (A) Agarose gel electrophoretic profile of the bacteriophage phiNY genome. (B, C) Transmission electron micrographs of the host bacterium, M. aerodenitrificans strain LH11-4, and bacteriophage phiNY. (D) The presence of double-stranded RNAs (dsRNAs) was detected from the phiNY-reinfected colony, but not the chloroform-treated phiNY-infected colony. (E) Growth curves of M. aerodenitrificans strain LH11-4 with phiNY (optical density at 595 nm [OD595], mean ± standard deviation [SD] of three biological replicates). (F) Quantification of phiNY by reverse transcription–quantitative real-time PCR (RT-qPCR) from the precipitate of M. aerodenitrificans strain LH11-4 (mean ± SD of three biological replicates). (G) Growth curves of M. aerodenitrificans strain LH11-4 with phiNY, phiNY-negative strain LH9, and the phiNY-reinfected strain LH9-NY (OD595, mean ± SD of three biological replicates). (H) Growth curves of M. aerodenitrificans strain LH11-4 with phiNY, phiNY-negative strain LH3, and the phiNY-reinfected strain LH3-NY (OD595, mean ± SD of three biological replicates).