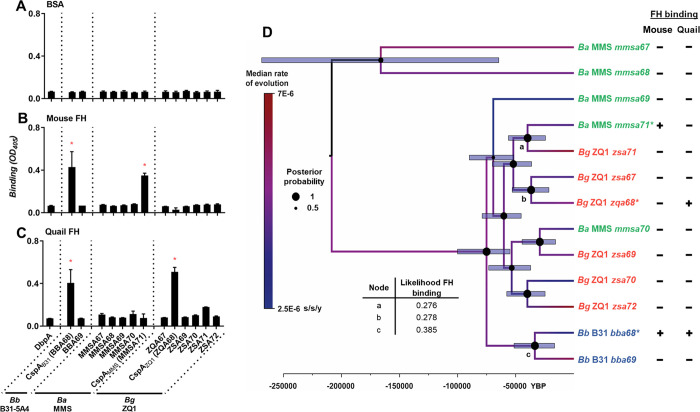
Fig 6. Allelically variable, host-specific FH-binding activity of CspA has emerged from convergent evolution.
(A to C) Two μM of histidine-tagged Pfam54-IV proteins from B. burgdorferi B31-5A4, B. afzelii PKo, or B. garinii ZQ1 or recombinant histidine-tagged DbpA from B. burgdorferi B31-5A4 (negative control) were added in triplicate to wells coated with of purified (A) BSA (negative control) or (B) mouse or (C) quail FH. Protein binding was measured by ELISA in three independent experiments. Each bar represents the geometric mean ± 95% confidence interval of three replicates in one representative experiment. Significant differences (p < 0.05, Kruskal-Wallis test with the two-stage step-up method of Benjamini, Krieger, and Yekutieli) in the levels of FH binding of indicated proteins relative to DbpA are indicated (*). The ability of each protein in binding to factor H is summarized in S1 Table. (D) Maximum clade credibility tree is based on Pfam54-IV genes from B. burgdorferi B31 (blue), B. afzelii MMS (green), B. garinii ZQ1 (red). The scale bar represents an approximate timeline of evolution, in years before present (“YBP”), using the estimated substitution rate of 4.75×10−6 substitutions/site/year. Node bars represent the 95% highest posterior density of the node age. Node circles represent the posterior probability support. Branches are colored based on estimated the median substitution rate as per the legend to the left. Maximum likelihood- and parsimony-based ancestral state reconstructions were used to estimate FH-binding activities at ancestral nodes.