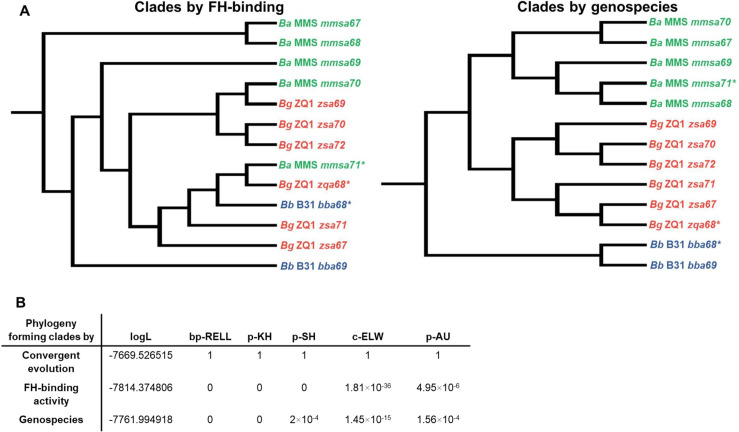
Fig 8. Competing evolutionary scenario evaluation of PFam54-IV gene divergence rejects alternative tree topologies of PFam54-IV proteins forming clades by FH-binding activity or spirochete genospecies.
(A) Phylogenetic scenarios showing a single emergence of FH-binding or PFam54-IV genes forming clades by genospecies. (B) Log-likelihood value for each tree (logL), RELL bootstrap proportion (bp-RELL; 1.0 is full support for that tree), Kishino-Hasegawa test, Shimodaira-Hasegawa test, Expected Likelihood Weights (c-ELW; 1.0 is full support for that tree), and the Approximately Unbiased test (p-AU). Significant p-values indicate comparisons where the best-known ML tree explains the sequence data better than the alternative scenario.