Figure 7.
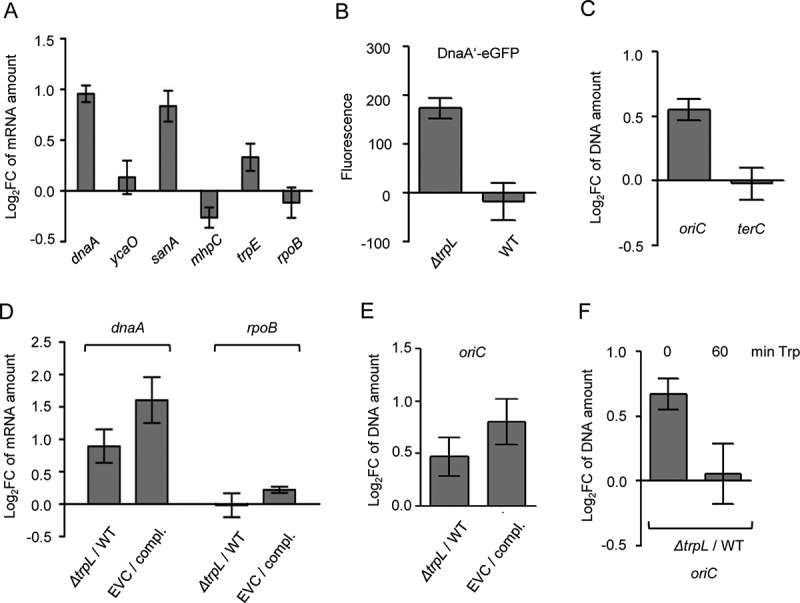
Comparison between the ΔtrpL mutant and the parental strain MG1655 confirms the role of Ec-rnTrpL in regulation of dnaA expression and initiation of chromosome replication. (A) The relative amount of the indicated mRNAs were analysed by qRT-PCR and the levels in the ΔtrpL mutant were compared to those in the parental strain MG1655. In vitro transcript was used as reference gene (spike in control). (B) Fluorescence of MG1655 ΔtrpL (pSRKGm-dnaA’-egfp) and MG1655 (pSRKGm-dnaA’-egfp) cultures supplemented with 0.2 mM IPTG. Normalization to EVC cultures was performed. (C) The levels of oriC and terC in the ΔtrpL mutant were compared to those in the parental strain using qPCR. Both strains contained pSRKGm, and the Gm resistance gene was used as reference gene. (D) The relative amounts of dnaA mRNA were analysed by qRT-PCR. The level in the EVC ΔtrpL (pSRKTc) was compared to the level in the complemented strain (compl.) ΔtrpL (pSRKTc-Ec-rnTrpL). Both strains were cultivated in the presence of 0.2 mM IPTG. In parallel, the level in the ΔtrpL mutant was compared to that in the wild type. The rpoB mRNA was analysed as a non-target RNA. A spike-in transcript was used as reference. (E) The oriC level was analysed by qPCR. The level in the EVC was compared to the level in the complemented strain (compl.) (for strain description and growth conditions, see panel D). In parallel, the ΔtrpL mutant was compared to the wild type. As a reference, terC was used. (F) The oriC level was analysed by qPCR. The level in the ΔtrpL mutant was compared to that in the wild type before and 1 h after addition of Trp (20 µg/ml). As a reference, terC was used. Cultures grown in minimal medium were used. Shown are means and standard deviations from three independent experiments
