Figure 2.
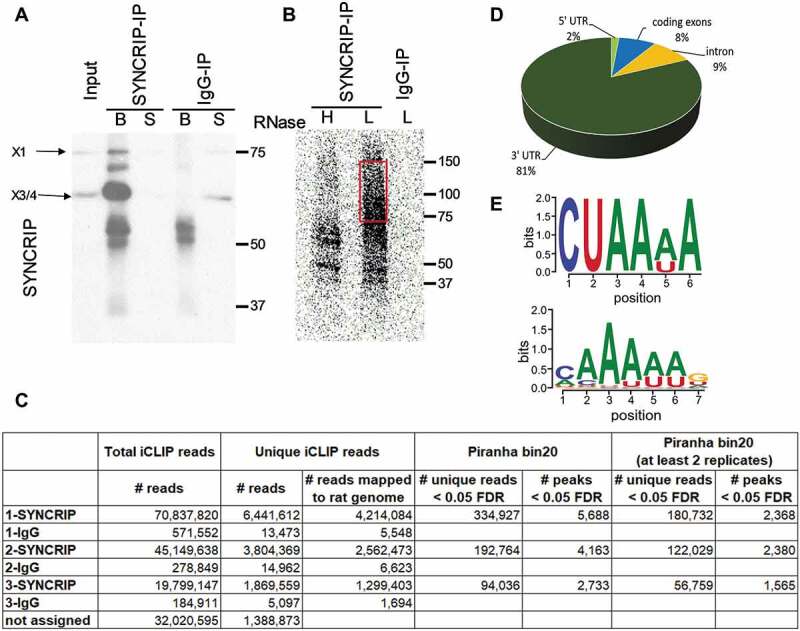
Identification of direct SYNCRIP cytoplasmic target mRNAs in rat cortical neurons using iCLIP. (A). Western blot of SYNCRIP or IgG-immunoprecipitation (IP) of rat cortical neuron extracts after cross-linking using anti-SYNCRIP antibodies (B: beads, S: supernatant). (B). Autoradiography of RNA transcripts cross-linked to SYNCRIP in high (H, 1:100 dilution) or low (L, 1:7000 dilution) RNase conditions. The migration of the X3/4 isoform is indicated. For downstream iCLIP applications cross-linked RNA transcripts corresponding to 70–150 kDa size (red box) are used. (C). Summary of identified unique and clustered (peaks) reads in each of the three SYNCRIP iCLIP replicates. Unique iCLIP reads are used as input for the peak-caller software Piranha to generate iCLIP peaks (clusters) with a bin_size 20 parameter (see materials and methods section). (D). Pie chart representing the distribution of identified iCLIP peaks (Piranha bin 20 – at least 2 replicates) in 5ʹUTR, coding sequence (CDS), intron and 3ʹUTR of pre-mRNAs. Total iCLIP peaks mapping to pre-mRNAs are taken as 100%. (E). SYNCRIP binding motifs. Predicted neuronal SYNCRIP binding motif (upper panel), as obtained by means of an unbiased motif search near iCLIP peaks of mRNA 3ʹUTRs against shuffled control sequences (E-value = 0.00062). SYNCRIP binding motif from CISBP-RNA database, based on large-scale in vitro analysis (33, lower panel)
