Figure 4.
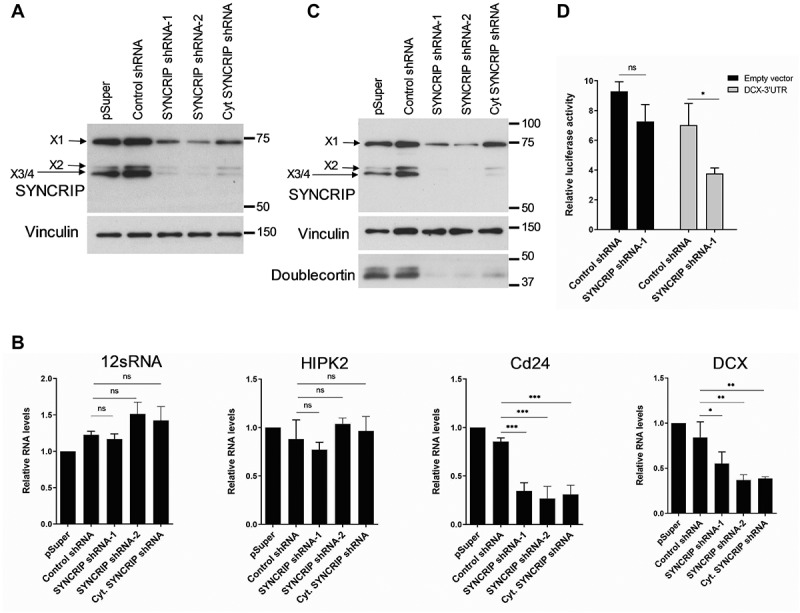
SYNCRIP positively regulates Dcx and Cd24 mRNA and protein expression. (A). Western blot using anti-SYNCRIP antibody in primary cortical neurons that had been nucleofected at DIV0 with the indicated shRNA constructs and lysed at DIV5. Vinculin was used as a loading control. (B). qPCR of the SYNCRIP iCLIP target mRNAs Cd24 and Dcx in neurons treated as in 4A (B).12sRNA and HIPK2 are used as negative controls. RNA levels were normalized to GAPDH mRNA. Data are presented as mean ± SD (n = 3). Statistical test is performed using one-way ANOVA (12sRNA: F (3,8) = 4.45; P = 0.041; HIPK2: F (3,8) = 2.23; P = 0.162; Cd24: F (3,8) = 27.15; P = 0.0002; DCX: F (3,8) = 11.48; P = 0.0029; Dunnett’s multiple comparisons test: adj. p value, * < 0.05, ** < 0.01, *** <0.001) (C). Western blot using the indicated antibodies with cell extracts from neurons treated as in 4A. A representative blot from a total of 3 blots is shown. (D). Luciferase reporter gene assay in rat cortical neurons (DIV 6–9) transfected with a pmirGLO-DCX-3ʹUTR and the indicated shRNA constructs. Transfection of an empty pmirGLO vector was used as a negative control. Relative luciferase activity (firefly/renilla activity) is presented as mean ± SD (n = 3). Statistical test is performed using two-way ANOVA (empty vector: P = n.s., DCX-3ʹUTR: P < 0.05; UTR: F (1,8) = 21.26, P = 0.0017; shRNA: F (1,8) = 25.48, P = 0.0010; UTR x shRNA: F (1,8) = 1.168, P = n.s.)
