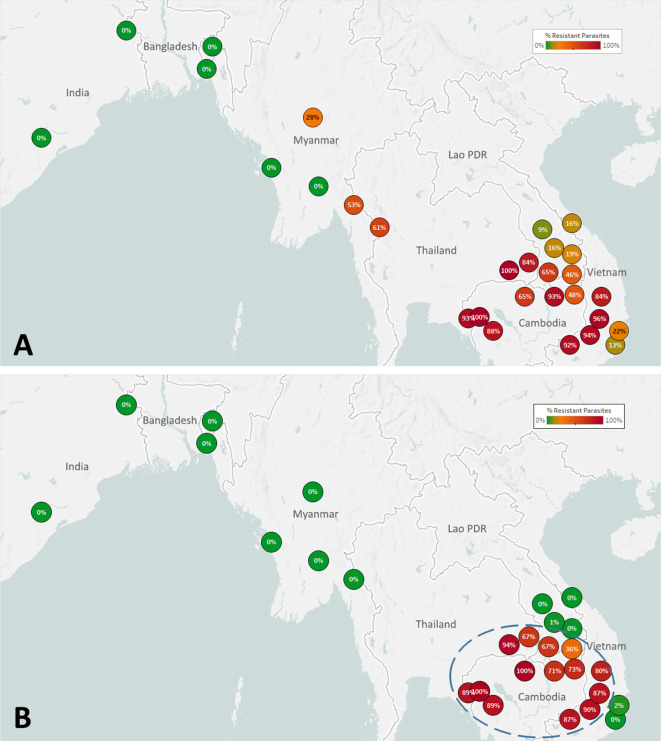
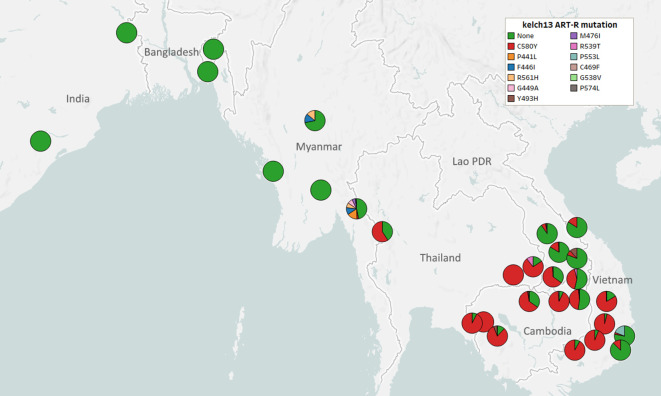
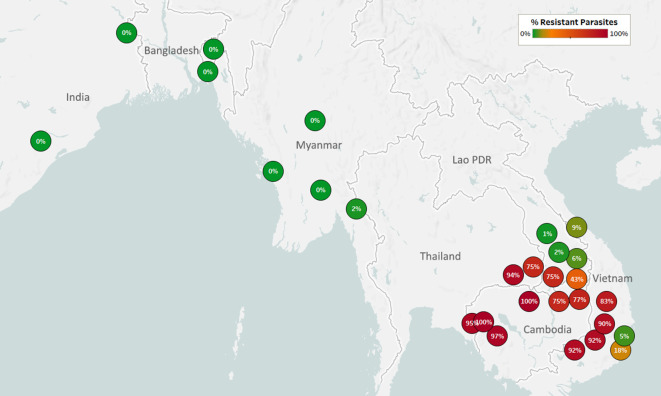
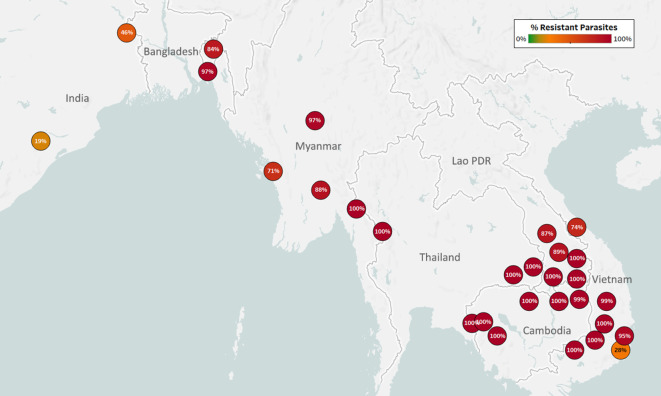
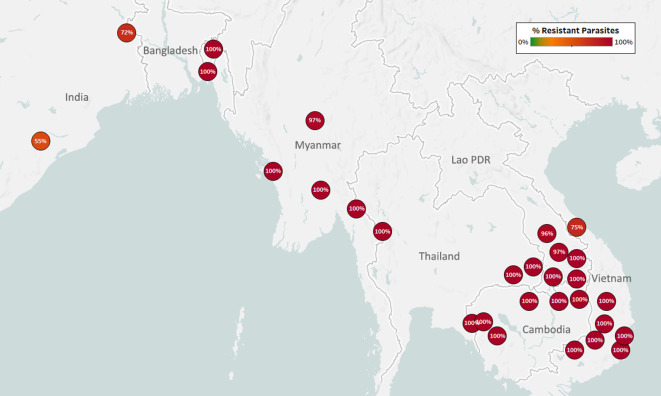
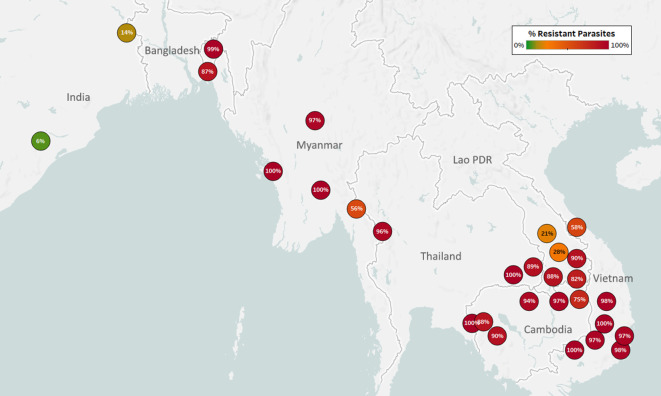
Figure 3. Map of the spread of (A) artemisinin resistance (ART-R) and (B) dihydroartemisinin-piperaquine resistance (DHA-PPQ-R) in Asian countries.
Marker text and color indicate the proportion of sample classified as resistant in each province/state/division surveyed. A total of 6762 samples were included in (A) and 3395 samples in (B), after excluding samples with undetermined phenotype prediction. The results are summarized in Table 3.