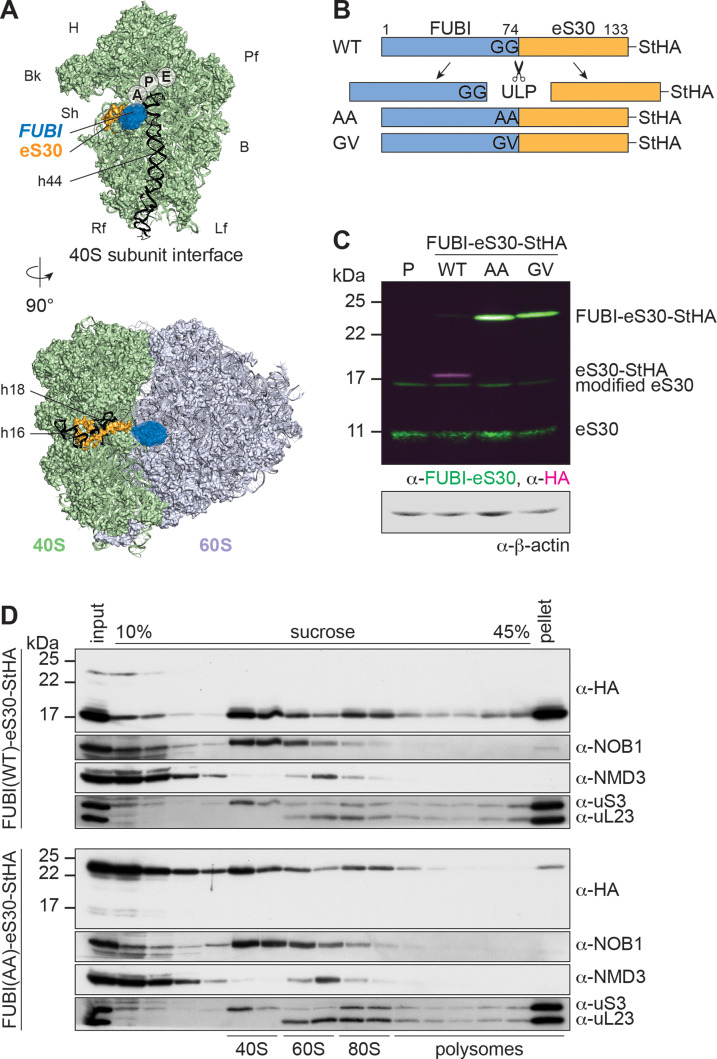
Figure 1. Mutant, non-cleavable FUBI-eS30 can be incorporated into (pre-)40S particles.
(A) Structures (PDB ID: 4UG0 Khatter et al., 2015) of the human 40S subunit shown from the subunit interface (top) and of the 80S ribosome viewed from the mRNA entry channel (bottom), highlighting the hypothetical location of FUBI (blue) at the N terminus of eS30 (orange). FUBI (PDB ID: 2L7R, state 1 (Lemak, 2011), marked with an opaque blue shape) was manually positioned in PyMOL avoiding molecular contacts in the surface representation mode. Ribosomal A-, P-, and E-sites are indicated and the 18S rRNA helices h44 (G1702–C1833), h18 (U595–A641), and h16 (C527–G558) are highlighted in black. B, body; Bk, beak; H, head; Lf, left foot; Pf, platform; Rf, right foot; Sh, shoulder. (B) Schematic representation of C-terminally StHA-tagged FUBI-eS30 wild-type (WT) and mutant G73,74A (AA) and G74V (GV) constructs used in this study. The C-terminal diglycine motif of FUBI (G73,G74) was mutated to impair FUBI removal by ubiquitin-like protease(s) (ULP). (C) Immunoblot of tetracycline-inducible HeLa cell lines expressing the indicated FUBI-eS30-StHA constructs using anti-FUBI-eS30/FAU and anti-HA primary antibodies. Fluorescent secondary antibodies against anti-FUBI-eS30/FAU (green) and anti-HA (magenta) antibodies were detected simultaneously. P, parental cells. Note that the StHA tag adds ~7 kDa to the (FUBI-)eS30 protein constructs. eS30 runs at a higher MW than the expected 7 kDa, likely due its high content in positively charged residues. (D) Extracts from FUBI-eS30-StHA WT (top) or AA mutant (bottom) HEK293 cell lines were separated on a linear 10–45% sucrose gradient by centrifugation. Expression of the FUBI-eS30-StHA constructs was induced with 0.1 µg/ml tetracycline for 17 hr. Input, gradient, and pellet fractions were analyzed by immunoblotting using the indicated antibodies.