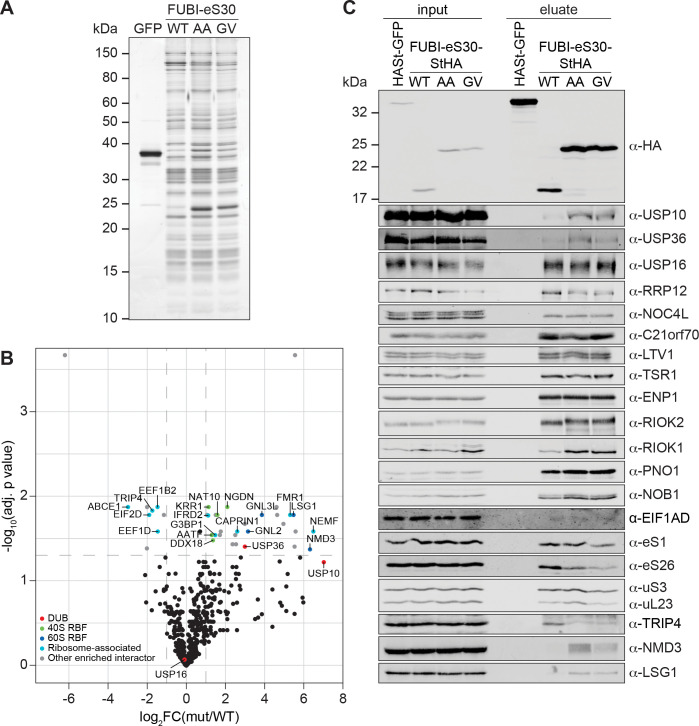
Figure 5. Identification of candidate proteases by differential affinity purification mass spectrometry of FUBI-eS30-StHA constructs.
(A) StrepTactin affinity purification of HASt-GFP (GFP), WT or mutant (AA, GV) FUBI-eS30-StHA from HEK293 cell lysates. Eluates were analyzed by SDS-PAGE followed by silver staining or mass spectrometry. (B) Results of the proteomic analysis of three biological replicates as in (A). The spectral counts of proteins that were compared to HASt-GFP confidently enriched on FUBI-eS30-StHA baits (SAINT Bayesian false discovery rate < 1%) were normalized to their size in amino acids. The log2 fold change of the average number of the spectral counts of the confident interactors identified on the non-cleavable AA and GV mutants (mut) vs. WT FUBI-eS30-StHA (log2FC(mut/WT)) are plotted against the negative log10 of the adjusted p value (-log10(adj. p value)). All confidently identified DUBs are labeled, significantly enriched interactors (|log2FC| > 1 and adj. p value < 0.05, demarcated with dashed lines) are categorized as indicated and individual proteins are labeled. Data before and after normalization and filtering are shown in Supplementary file 1. (C) Inputs and eluates of StrepTactin affinity purification performed as in (A) were analyzed by immunoblotting using the indicated antibodies.