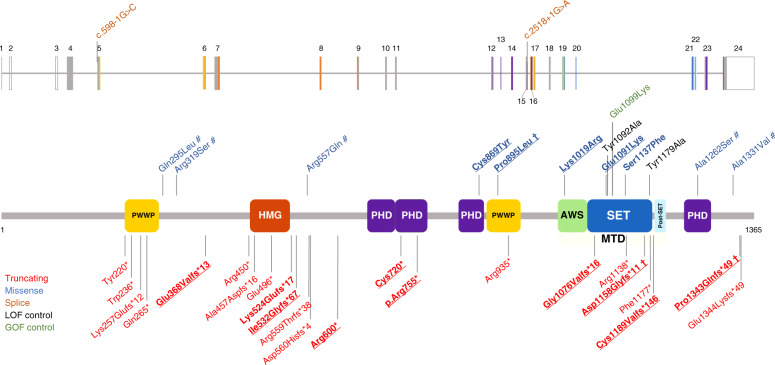
Fig. 1. Localization of the NSD2 variants.
The diagram shows the structure of the NSD2 gene (above) and protein (below, isoform 1, encoded by the NM_133330.2 transcript) together with the variants discussed in this study as well as variants that have previously been reported in large sequencing studies. The variants carried by the 18 additional individuals described in this study are in bold. Observed germline variants absent in the HGMD database are underlined. † Shared by more than one patient; # variant of uncertain significance (VUS)/likely benign. The observed pathogenic missense variants we found map to three distinct domains of NSD2: a PHD zinc-finger domain (residues 831–875), a PWWP domain (residues 880–942), and the catalytic methyltransferase domain (residues 1011–1203; composed of three subdomains, namely an AWS, a SET, and a post-SET domain). The color coding of the variants and protein domains is reported in the legend. AWS Associated With SET domain (IPR006560), GOF gain of function, HMG high mobility group box domain (IPR009071), LOF loss of function, MTD catalytic methyltransferase domain, composed by the AWS, SET and a post-SET domain, PHD zinc-finger domain, Plant-HomeoDomain type (IPR001965), PWWP proline–tryptophan–tryptophan–proline domain (IPR000313), SET Su(Var)3-9, enhancer-of-zeste, trithorax domain (IPR001214). Domains were annotated according to the Uniprot databank (see Web Resources).