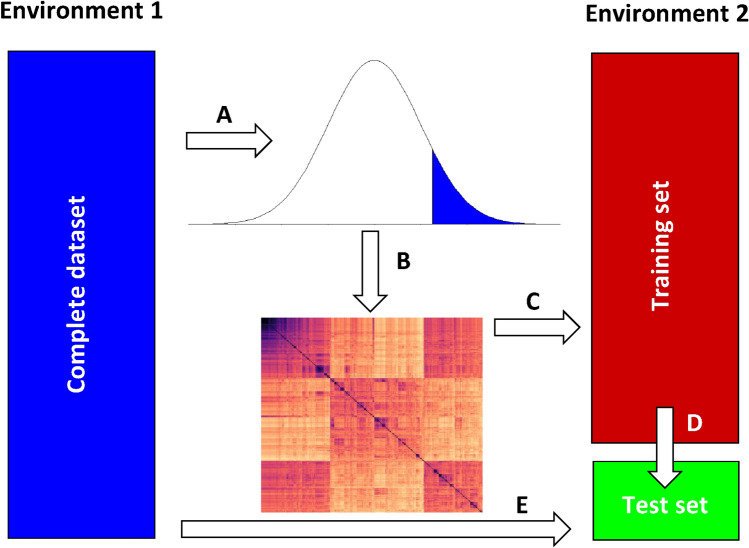
Fig. 1.
Basic scheme of uni- and bivariate sERRBLUP across environments. All pairwise SNP interaction effects and their variances are estimated from all data in environment 1, and effects are ordered either by absolute effect size or effect variance (A). Then, an epistatic relationship matrix for all lines is constructed from the top ranked subset of interaction effects (B) which in the univariate model is used in environment 2 (C) to predict phenotypes of the test set (green) from the respective training set (red, D). In the bivariate model, this information is combined with the complete data from environment 1 (blue, E) to predict the test set