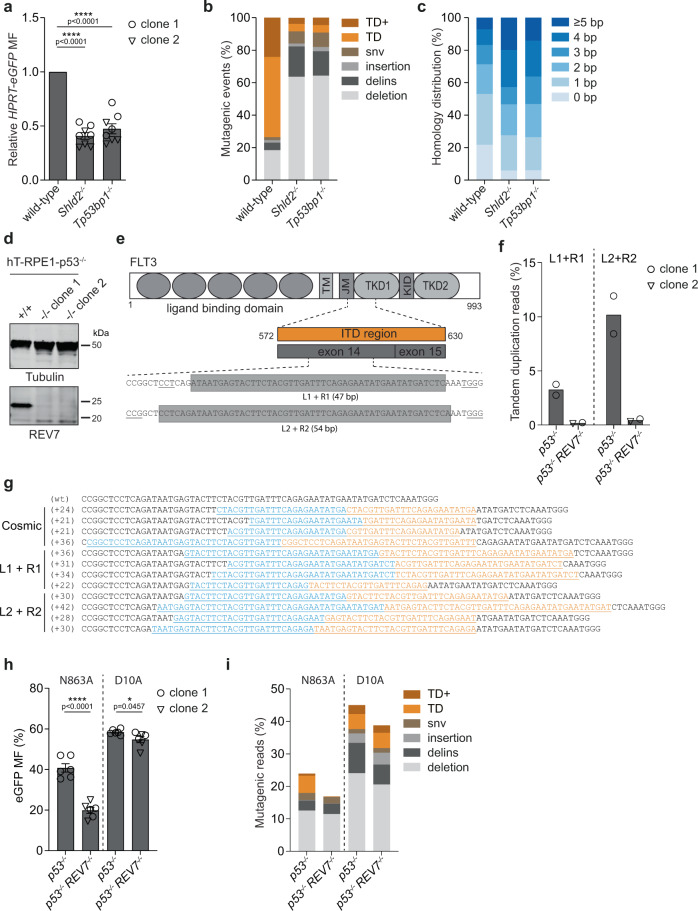
Fig. 3. A requirement for the 53BP1-Shieldin axis in TD formation in human and mouse cells.
a Relative HPRT-eGFP mutation frequency for the indicated mES cell lines (using two independent clones per genotype) transfected with Cas9‐N863A and sgRNAs targeting eGFP, producing DSBs with 3′ ssDNA overhangs. The data shown represent the mean ± SEM (n = 6) and are expressed as a fraction of the mutation frequency observed in wild‐type cells (set to 1). Statistical significance was calculated via a two-tailed unpaired t‐test. ****P ≤ 0.0001. b Quantification of the types of mutagenic events at breaks with 3′ ssDNA overhangs for the indicated mES cell-lines transfected with Cas9-N863A and sgRNAs targeting eGFP. GFP-negative (mutant) cells were sorted and used for targeted sequencing around the break-site, data represent the average of two independent clones per genotype. c Quantification of the extent of microhomology used for deletions and tandem duplications (TD) in the indicated genotypes. d Immunoblot confirming the loss of REV7 protein expression in two independent RPE1-p53−/− REV7 knockout clones (lower panel). An immunoblot for Tubulin is included as a loading control (upper panel). A representative example of at least two independent immunoblots is shown. e Targeting strategy of the human FLT3 gene. The internal tandem duplication (ITD) region in exon 14 of the human FLT3 gene was targeted using Cas9-N863A and two separate combinations of sgRNAs: L1 + R1 and L2 + R2, resulting in DSBs with 3′ overhangs of 47 bp and 54 bp, respectively; PAM-sites are underlined. f Quantification of the number of TD and TD+ reads derived from human RPE1-p53−/− and RPE1-p53−/− REV7−/− cells electroporated with Cas9-N863A and the indicated sgRNAs. Unselected cells were used for targeted sequencing around the break site. Data represents the average of RPE1-p53−/− samples and independent RPE1-p53−/− REV7−/− clones (n = 2). g Sequence representation of tandem duplication in the human FLT3 gene found in the Cosmic database (top) and in RPE1-p53−/− cells transfected with Cas9-N863A and the indicated sgRNAs (bottom). The sequence that is duplicated is underlined in cyan and orange. h absolute eGFP mutation frequency for RPE1-p53−/− and RPE1-p53−/− REV7−/− cells stably expressing Cas9-N863A or Cas9-D10A transfected with sgRNAs targeting eGFP. The data shown represent the mean ± SEM (n = 6). Statistical significance was calculated via a two-tailed unpaired t‐test. *P ≤ 0.05, ****P ≤ 0.0001. i Quantification of the number of mutagenic reads at breaks with 3′ ssDNA or 5′ ssDNA overhangs for the indicated RPE1 cell-lines expressing Cas9-N863A or Cas9-D10A, respectively, and transfected with sgRNAs targeting eGFP. Unselected cells were used for targeted sequencing around the break-site, data represent the average of four independent samples per genotype. MF, mutation frequency; Delins, deletion insertion; snv, single nucleotide variant; TD, tandem duplication; TD+, tandem duplication with additional mutation (see “Methods” section); bp, base pair.