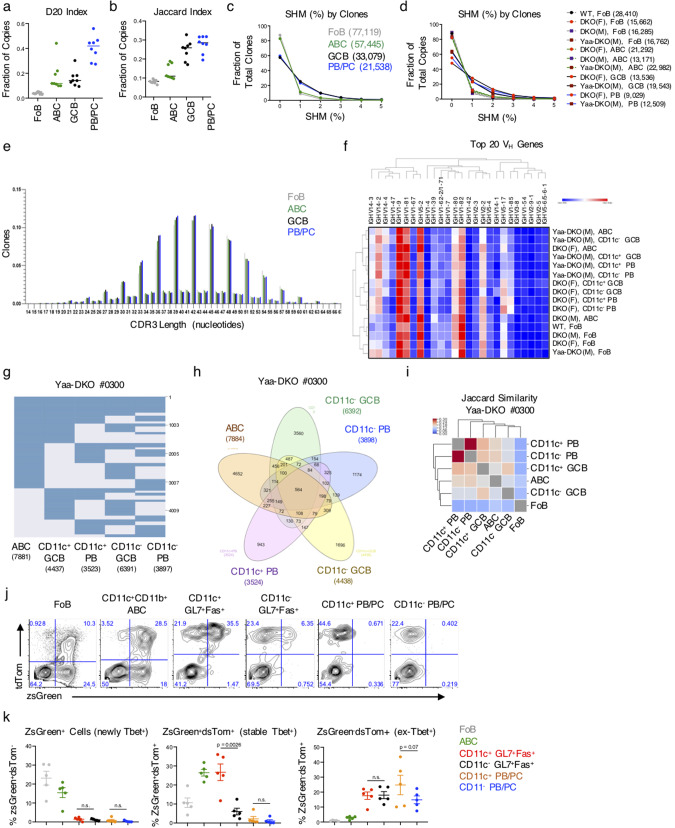
Fig. 3. B cell effector populations exhibit a high degree of interrelatedness.
IgH sequencing was performed on sorted FoBs (B220+CD11c−CD11b−CD23+) (gray circles), ABCs (B220+CD11c+CD11b+) (green circles), CD11c+ and CD11c− GL7+CD38lo B cells (gated on B220+ splenocytes) (black circles), and CD11c+ and CD11c− PB/PCs (B220loCD138+) (blue circles) from aged (24+wk) female C57BL/6 (WT), female DKO(F), male DKO(M), and male Yaa-DKO(M) mice. a D20 index of each population aggregated across all strains. D20 indicates the number of total copies in the top 20 clones as a fraction of total copies across all clones. Data show mean with each dot representing a single mouse; n = 8 mice. b Plot showing the level of overlap between clones in different sequencing libraries prepared from the same sample aggregated across all strains. For each comparison, each clone was only counted once (no weighting for clone size) and functional overlap was computed using the Jaccard Index. Data show mean with each dot representing a single mouse; n = 8 mice. c Plot showing somatic hypermutation (SHM) as a fraction of total clones for each subset aggregated across mouse strains. The numbers in parenthesis indicate the total number of clones in the given subset. Each clone counts once for each subset. If a clone overlaps in multiple subsets, the SHM for each subset was calculated just for the sequences in a given subset. d The SHM by clones is shown across each mouse strain/subset combination as in Fig. 3c. e Plot showing CDR3 length (in nucleotides) of clones aggregated across all strains. Each clone is counted once in each subset/strain combination; n = 8. f Heatmap showing usage of the 20 most frequent VH genes. Each clone is counted only once. Data are normalized by row and visualized in Morpheus using the default settings. g Plot showing clones (rows) that overlap between at least two B cell subsets (columns). Numbers along the right side of the plot indicate clone counts. Data from a single analysis of subsets from a Yaa-DKO mouse. h Venn diagram showing clonal overlap where numbers indicate clone counts in the different subset interactions. Data from a single analysis of subsets from a Yaa-DKO mouse. i Heatmap showing Jaccard similarity (fraction of clones that overlap between different two-subset comparisons). Diagonal values are excluded for scaling. Data from a single analysis of subsets from a Yaa-DKO mouse. j, k Female ZTCE-DKO mice were treated with tamoxifen to mark Tbet-expressing cells with tdTomato expression for 3d. Representative FACS plots (j) and quantifications (k) of zsGreen and tdTomato expression from the indicated populations. Data show mean ± SEM; n = 5 over 2 independent experiments; p-value by paired two-tailed t-tests.