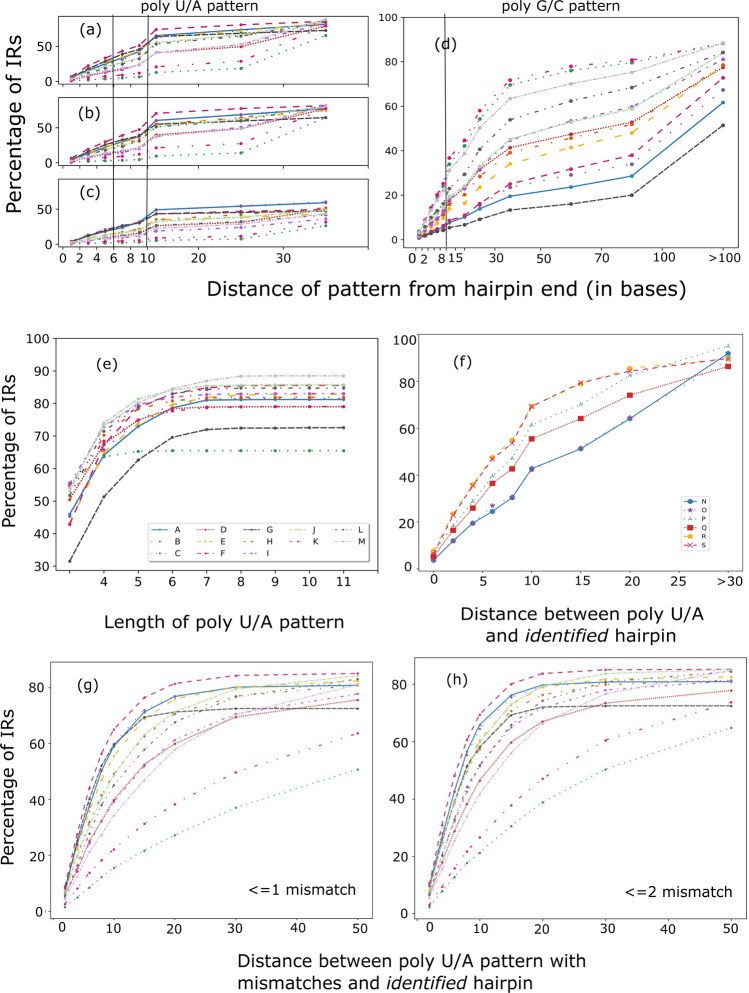
Figure 8.
Line plots showing trends of poly U/A and G/C pattern occurrence in IRs. (a–c) Plots show the distribution of poly A/U patterns at a given distance between the hairpin end, where the first occurring poly U/A pattern has been considered. (a) depicts the statistics for all IRs where we have identified hairpins, (b) cluster + single RNA-seq derived hairpin units, (c) single RNA-seq derived hairpin units. Plot (d) shows the distance between identified hairpin units and the first occurring poly G/C pattern. Vertical lines at distances 6, 10 in (a–c), and only 10 in (d) are drawn to highlight the low occurrence of patterns close to the hairpin end. Plot (e) shows the lengths of the first A/U patterns in all IRs. The different colors from (a–e) and in (g), (h) are the 13 bacterial genomes analyzed and follow the same order as in Table 1 and shown in legend with the plot (e). Plot (f) shows the distance between the hairpin and the first occurring pattern after them for identified hairpin units where genomes are N: Escherichia coli K-12 substr MG1655, O: Escherichia coli O127-H6 str. E2348, P: Bacillus subtilis subsp. subtilis delta6, Q: Bacillus subtilis subsp. subtilis N3-1, R: Helicobacter pylori PeCan4, S: Helicobacter pylori Lithuania75. Plots (g) and (h) show the same statistics as (a) where the poly A/U patterns have ≤ 1 or ≤ 2 mismatches, respectively.