Abstract
During 2020, understanding the molecular mechanism of SARS-CoV-2 infection (the cause of COVID-19) became a scientific priority due to the devastating effects of the COVID-19. Many researchers have studied the effect of this viral infection on lung epithelial transcriptomes and deposited data in public repositories. Comprehensive analysis of such data could pave the way for development of efficient vaccines and effective drugs. In the current study, we obtained high-throughput gene expression data associated with human lung epithelial cells infected with respiratory viruses such as SARS-CoV-2, SARS, H1N1, avian influenza, rhinovirus and Dhori, then performed comparative transcriptome analysis to identify SARS-CoV-2 exclusive genes. The analysis yielded seven SARS-CoV-2 specific genes including CSF2 [GM-CSF] (colony-stimulating factor 2) and calcium-binding proteins (such as S100A8 and S100A9), which are known to be involved in respiratory diseases. The analyses showed that genes involved in inflammation are commonly altered by infection of SARS-CoV-2 and influenza viruses. Furthermore, results of protein–protein interaction analyses were consistent with a functional role of CSF2 and S100A9 in COVID-19 disease. In conclusion, our analysis revealed cellular genes associated with SARS-CoV-2 infection of the human lung epithelium; these are potential therapeutic targets.
Subject terms: Computational biology and bioinformatics, Immunology, Diseases
Introduction
Infection of severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2) is the cause of human coronavirus disease 2019 (COVID-19). The recent pandemic has caused devastation due to rapid spread of this viral infection. As a respiratory illness, the disease is readily transmitted. It also has a long incubation and can be carried asymptomatically, thus spreading through communities1. The COVID-19 pandemic has affected almost every country, regardless of their medical infrastructure and economic status. It has caused a healthcare crisis and created a devastating economic burden, including high unemployment, which exacerbates the effect of the disease2. At present, more than 70 million people from 213 countries have been infected with the virus (https://www.worldometers.info/coronavirus/). The rapid spreading of this respiratory infection has forced millions to shelter in their homes and has led to death of more than 1,580,000 individuals. Additionally, in the U.S., COVID-19 has disproportionately affected patients, particularly minorities and those with chronic problems such as hypertension, lung disease, diabetes, and immunocompromised conditions.
In the last three decades, the world has witnessed zoonotic transmission of various viruses (from animals to humans) leading to severe respiratory complications. These include H1N1, avian influenza, severe acute respiratory syndrome (SARS), and Middle East respiratory syndrome coronavirus (MERS‐CoV)3,4. Although infections of these viruses are often fatal, their effect is generally restricted to geographic locations such as Africa, Asia, and South America. As the world awaits a vaccine for SARS-CoV-2, efforts are being made to understand the molecular mechanisms of these infections5–9.
High-throughput technologies such as RNA sequencing and microarrays are useful in the detection of respiratory virus infections and in understanding their molecular effect on human lung epithelial cells10. Extensive data on corona virus sequences have been deposited in public repositories such as NCBI Gene Expression Omnibus (GEO) and EMBL Array Express11,12. Meta-analysis and mining of such data can aid in a) understanding the molecular impact of COVID-19, b) elucidating differences and similarities between SARS-CoV-2 and other respiratory virus infections, and c) identifying targets for drug development. In the current study, we performed comparative analysis of publicly available gene expression data related to human lung epithelial cells infected with a respiratory virus. The analyses identified genes specifically expressed by SARS-CoV-2 infections and those that are commonly altered due to infection of coronovirus-2 and/or other respiratory viruses. In particular, expression of CSF2 (colony-stimulating factor 2) appears to be involved in COVID-19 disease. Several COVID-19 clinical trials are currently focusing on inhibition of CSF2 [GM-CSF]13–15.
Results
RNA sequencing identified genes altered on SARS-CoV-2 infection of normal human lung epithelial cells
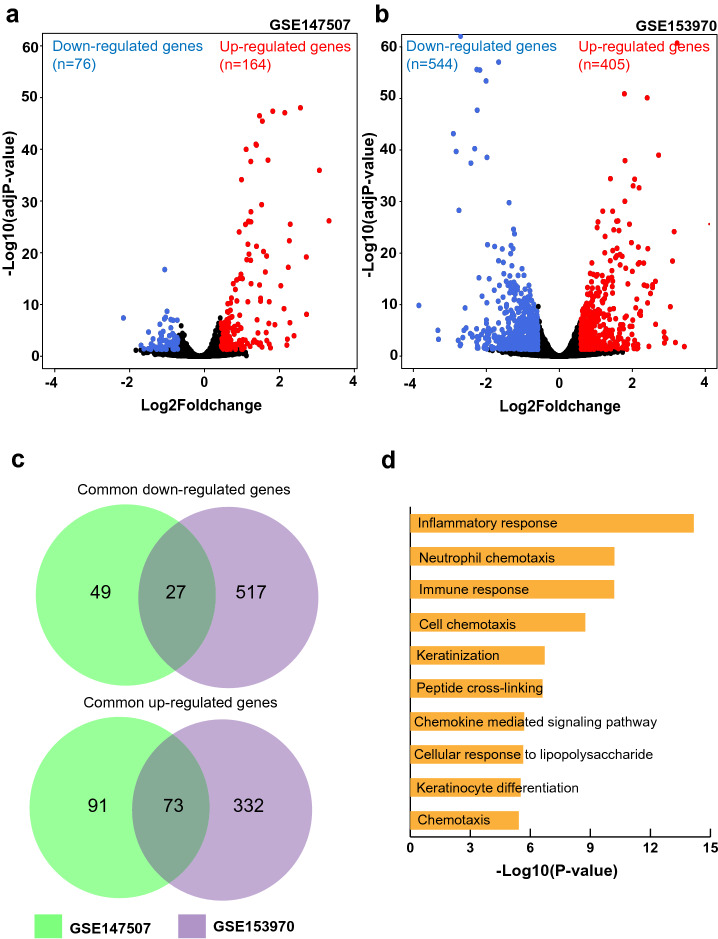
Differential expression analysis was conducted separately using RNA-seq data from GSE147507 and GSE153970. In total, 164 genes were up-regulated, and 76 genes were down-regulated on SARS-CoV-2 infection of normal human bronchial epithelial cells [GSE147507] (Fig. 1a). Similarly, 405 genes were up-regulated and 544 genes were down-regulated in SARS-CoV-2-infected human airway epithelium [GSE153970] (Fig. 1b). Gene list comparison identified 27 commonly down-regulated and 73 commonly up-regulated genes (Fig. 1c). Gene Ontology enrichment analysis of common differentially expressed genes showed “Inflammatory response”, “Neutrophil chemotaxis”, “Immune response”, “Cell chemotaxis”, and “Keratinization” as the top 5 enriched biological processes (Fig. 1d).
Figure 1.
High throughput gene expression analysis of SARS-CoV-2 infection in human lung epithelial cells. (a,b) Volcano plots showing the proportion of differentially expressed genes on SARS-CoV-2 infection in human lung epithelial cells [from GSE147507 and GSE153970, respectively]. (c) Venn diagram depicting genes commonly up-/down-regulated as per both studies. (d) Top 10 biological processes associated with common differentially expressed genes on SARS-CoV-2 infection of human lung epithelial cells.
The impact of other viral infections on the human lung epithelial cell transcriptome was explored using publicly available microarray data
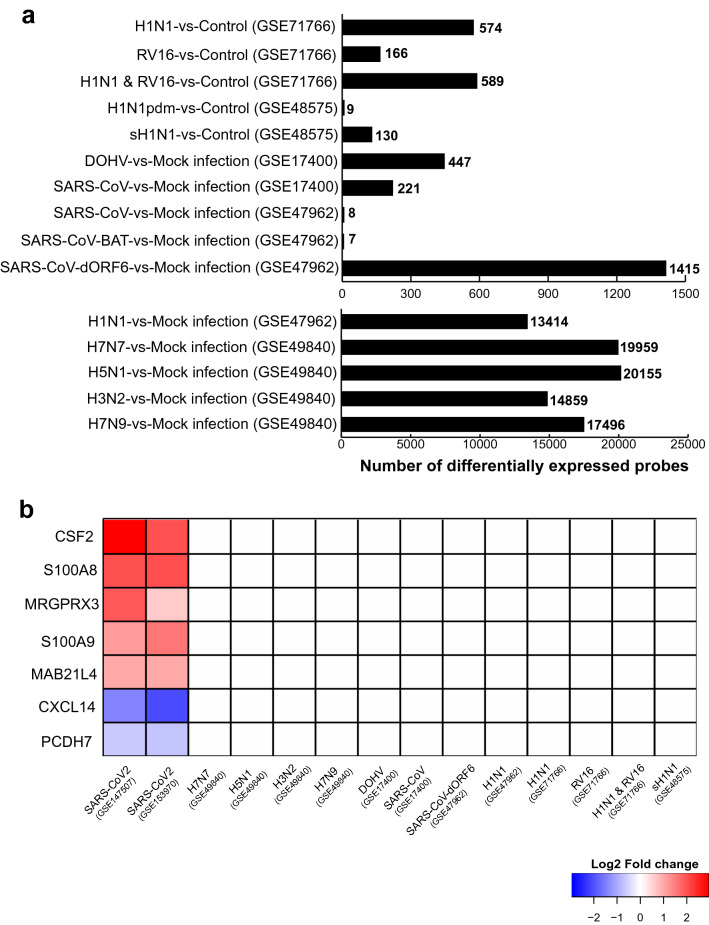
A search of microarray datasets in the NCBI GEO database led to identification of five studies involving human lung epithelial cells subjected to viral infection. GEO2R analyses of each study were performed separately to identify differentially expressed genes. Figure 2a shows differentially expressed probes identified from each comparative analysis. From GSE71766, there were 574 differentially expressed probes (281 genes) in H1N1-infected BEAS-2B cells compared to control; 166 probes (72 genes) were up-regulated by RV16 infection. The combined infection of RV16 and H1N1 altered expression of 589 probes (288 genes). In the case of GSE49840, there were 19,959 probes (14,892 genes) differentially expressed in H7N7-infected Calu-3 cells compared to mock infected cells; 20,155 (14,948 genes), 14,859 (11,362 genes), and 17,496 probes (13,012 genes) were differentially expressed by H5N1, H3N2, or H7N9 infections, respectively. For GSE17400, DOHV or SARS-CoV infection of Calu-3 cells led to altered expression of 447 (344 genes) and 221 probes (182 genes), respectively. Analysis of GSE48575 led to identification of 130 (99 genes) and 9 (7 genes) differentially expressed probes on seasonal (sH1N1) or pandemic (H1N1pdm) influenza virus infection, respectively, of NHBECs. Lastly, the processing of GSE47962 resulted in discovery of 13,414 (7938 genes), 1415 (997 genes), 7 (5 genes), and 8 (6 genes) differentially expressed probes after H1N1, SARS-CoV-dORF6, SARS-CoV-BAT, or SARS-CoV viral infections, respectively, of HAE cells.
Figure 2.
Comparative transcriptome analysis of collected gene expression data. (a) Histogram showing the differentially expressed genes as determined by GEO2R analysis of respiratory viral infection microarray data. (b) Heatmap showing 7 genes exclusively altered on SARS-CoV-2 infection.
Comparative analysis of DEGs resulted in identification of SARS-CoV-2 infection-specific genes and those commonly affected by most of the viral infections
In order to identify genes that are exclusively affected on SARS-CoV-2 infection, we compared common DEGs (98 protein-coding genes of 100) from RNA-seq studies with GEO2R analysis results of GSE47962, GSE17400, GSE48575, GSE49840, and GSE71766 (Supplementary Table S1). GEO2R results from each microarray study include lists of differentially expressed probes, gene symbols, fold change, and adjusted p-values. If multiple probes related to the same gene were differentially expressed, we considered the probe with highest fold change value. Comparative analysis showed 7 genes exclusively altered on SARS-CoV-2 infection, including 5 up-regulated genes (CSF2, S100A8, MRGPRX3, S100A9 and MAB21L4) and two down-regulated genes (CXCL14 and PCDH7) (Fig. 2b). However, microarray platforms showed the absence of probes related to MAB21L4 in all microarray studies (Table 1).
Table 1.
List of probes associated with COVID-19 exclusive genes in microarray platforms related to selected gene expression studies.
| Gene | Affymetrix human genome U133 plus 2.0 array | Affymetrix human genome U219 array | Agilent-014850 whole human genome microarray 4 × 44 K G4112F | Agilent-039494 SurePrint G3 human GE v2 8 × 60 K microarray | Illumina Human HT-12 V4.0 expression beadchip |
|---|---|---|---|---|---|
| CSF2 |
210228_at 210229_s_at |
11728876_at | A_23_P133408 | A_23_P133408 | ILMN_1661861 |
| S100A9 | 203535_at | 11716523_at | A_23_P23048 | A_23_P23048 | ILMN_1750974 |
| S100A8 | 214370_at | 11753823_a_at | A_23_P434809 | A_23_P434809 | ILMN_1729801 |
| MAB21L4 | |||||
| MRGPRX3 | 1553293_at | 11738052_at | A_23_P389371 | ILMN_1773546 | |
| CXCL14 | 218002_s_at |
11756059_a_at 11717910_at 11717911_x_at 11717912_s_at |
A_23_P213745 | ILMN_1748323 | |
| PCDH7 |
228640_at 205534_at 205535_s_at |
11737384_at 11741722_at 11724981_at 11724982_s_at 11724983_at |
A_23_P212888 A_23_P378364 A_24_P914638 A_23_P310921 |
A_33_P3509019 | ILMN_1766668 |
Inflammation-related genes (such as IL6, IL1A, IL1B, CXCL2, CXCL6, CCL20, TNIP1, VNN1, TNFAIP3 and NFKBIZ) were commonly affected due to infection of SARS-CoV-2 and other SARS or avian/human influenza viruses (Supplementary Figure S1c).
Validation of SARS-CoV-2 exclusive genes in human bronchial organoid RNA-seq data
Processing and analysis of RNA-seq data related to SARS-CoV-2-infected human bronchial organoids (HBO) led to identification of 1532 differential expressed genes (861 up-regulated and 671 down-regulated genes) (Supplementary Figure S1a). With the exception of MAB21L4, all SARS-CoV-2 exclusive genes from comparative transcriptome analysis showed the same expression pattern in HBO cells (Supplementary Figure S1b).
Protein–protein interaction analysis of altered genes on SARS-CoV-2 infection revealed hub genes
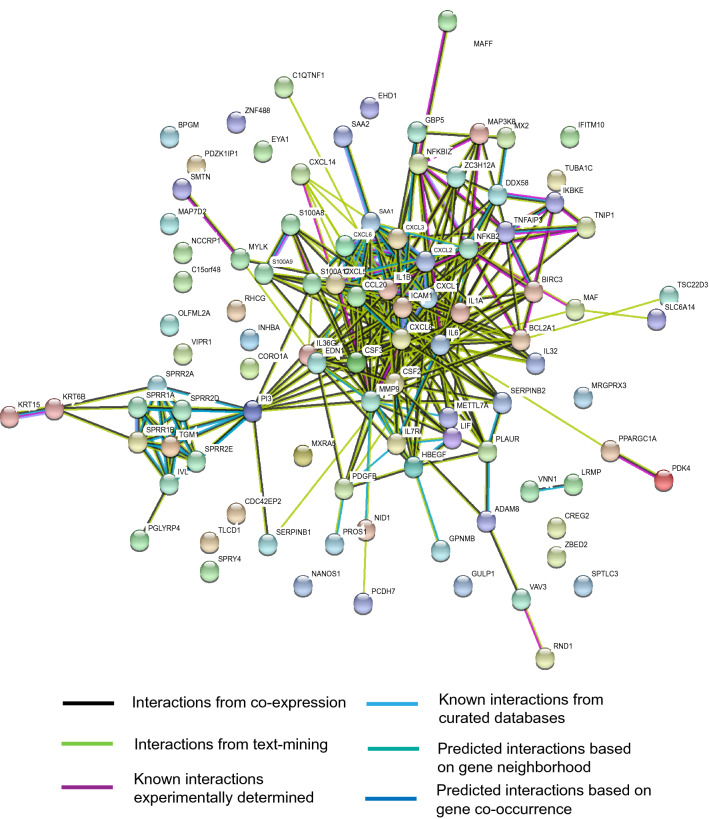
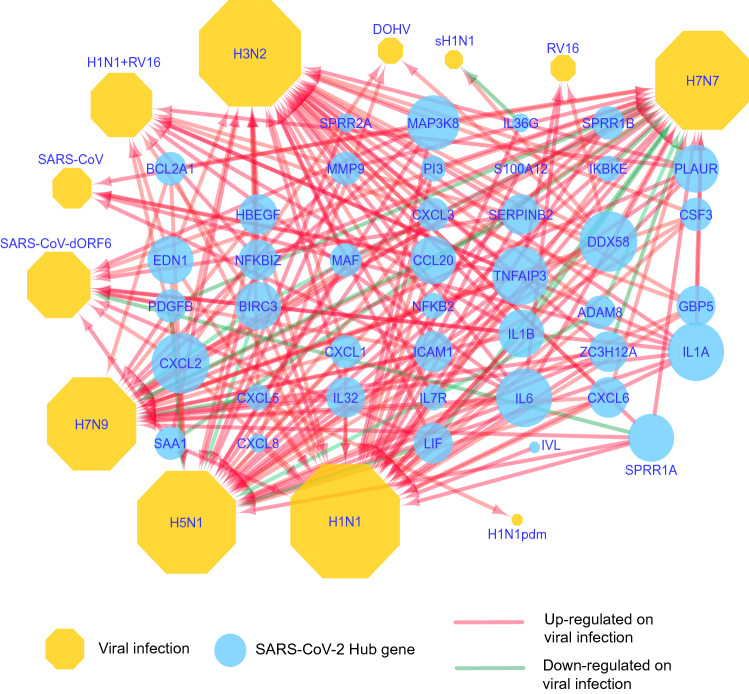
The common 98 protein-coding, differentially expressed genes on SARS-CoV-2 infection were first queried in the STRING database to identify known/predicted interactions among them. The database returned a PPI network of 333 interactions (edges) between 72 genes (nodes) (Fig. 3). The PPI network was downloaded as a simple interaction format (SIF) file, visualized with Cytoscape, and analyzed with Cytohubba plugin to identify hub genes. The top 50 genes were obtained based on three network parameters: degree, closeness, and betweenness separately. The 43 genes featured in all three lists were considered as hub genes (Table 2). The hub genes were IL6, IL1B, CXCL8, MMP9, CXCL1, CSF2, CCL20, ICAM1, IL1A, CXCL2, CSF3, SAA1, NFKB2, PI3, TNFAIP3, CXCL3, CXCL5, CXCL6, EDN1, HBEGF, BCL2A1, NFKBIZ, S100A12, PLAUR, BIRC3, IL36G, LIF, SERPINB2, SPRR1A, SPRR1B, IVL, DDX58, SPRR2A, ZC3H12A, S100A9, PDGFB, IL7R, MAP3K8, IKBKE, MAF, ADAM8 and GBP5. Among hub genes, most were affected by infection of human or avian influenza viruses such as H7N7, H1N1, H7N9, H3N2, and H5N1 (Fig. 4); CSF2 and S100A9 were exclusive to SARS-CoV-2 infection.
Figure 3.
Protein–protein interaction network of genes altered by SARS-CoV-2 infection. The network with 333 edges among 72 nodes was obtained from the STRING database. Interactions with scores of ≥ 0.4 were considered.
Table 2.
Hub genes involved in SARS-CoV-2 infection of normal human bronchial epithelial cells.
| Hub nodes | Degree | Closeness | Betweenness | GSE147507 | GSE153970 | ||
|---|---|---|---|---|---|---|---|
| Absolute FC | P-value | Absolute FC | P-value | ||||
| IL6 | 39 | 52.66 | 1061.909 | 7.59 | 6.59E−20 | 5.87 | 4.78E−14 |
| IL1B | 35 | 50.16 | 715.3514 | 2.11 | 9.62E−25 | 1.67 | 5.66E−11 |
| CXCL8 | 34 | 49.66 | 382.1511 | 5.28 | 2.6E−112 | 2.38 | 5.98E−24 |
| MMP9 | 30 | 47.91 | 832.9246 | 5.50 | 4.86E−23 | 2.41 | 1.51E−14 |
| CXCL1 | 26 | 43.95 | 143.7859 | 2.64 | 2.27E−38 | 2.63 | 3.04E−19 |
| CSF2 | 23 | 43.83 | 194.193 | 7.63 | 8.01E−09 | 3.97 | 6.06E−05 |
| CCL20 | 22 | 43.16 | 99.528 | 9.31 | 1.37E−71 | 3.44 | 1.25E−51 |
| ICAM1 | 21 | 42.83 | 130.012 | 3.66 | 1.24E−38 | 1.53 | 6.44E−05 |
| IL1A | 19 | 40.28 | 23.204 | 2.14 | 8.95E−16 | 1.71 | 5.09E−10 |
| CXCL2 | 19 | 40.45 | 39.909 | 2.65 | 9.33E−15 | 2.15 | 1.75E−09 |
| CSF3 | 18 | 39.61 | 18.700 | 32.18 | 7.74E−18 | 9.36 | 1.77E−61 |
| SAA1 | 17 | 38.95 | 142.325 | 5.04 | 8.62E−48 | 1.56 | 1.53E−06 |
| NFKB2 | 17 | 39.61 | 122.589 | 2.01 | 1.82E−15 | 1.78 | 3.29E−08 |
| PI3 | 17 | 40.75 | 1217.573 | 3.72 | 4.64E−07 | 1.93 | 2.59E−07 |
| TNFAIP3 | 17 | 38.56 | 46.582 | 3.11 | 3.38E−47 | 1.73 | 1.11E−08 |
| CXCL3 | 17 | 39.28 | 22.292 | 4.67 | 2.38E−14 | 3.67 | 9.64E−15 |
| CXCL5 | 15 | 38.11 | 8.458 | 11.70 | 6.87E−27 | 4.18 | 4.61E−35 |
| CXCL6 | 14 | 37.61 | 7.723 | 3.26 | 0.016423 | 2.74 | 5.63E−13 |
| EDN1 | 13 | 39 | 122.702 | 2.13 | 1.19E−08 | 1.88 | 1.94E−10 |
| HBEGF | 11 | 35.95 | 144.650 | 2.52 | 8.02E−27 | 2.75 | 2.78E−15 |
| BCL2A1 | 11 | 36.11 | 139.405 | 4.97 | 6.48E−10 | 3.30 | 4.38E−20 |
| NFKBIZ | 11 | 35.4 | 139.988 | 1.82 | 5.76E−12 | 2.28 | 7.37E−29 |
| S100A12 | 11 | 37.5 | 25.471 | 2.21 | 0.0438 | 5.28 | 2.45E−14 |
| PLAUR | 9 | 35.2 | 74.008 | 1.65 | 2.5E−06 | 2.25 | 2.30E−16 |
| BIRC3 | 9 | 34.53 | 8.979 | 3.36 | 5.7E−21 | 1.60 | 2.55E−05 |
| IL36G | 9 | 34.95 | 0.821 | 6.79 | 9.54E−49 | 14.64 | 1.43E−88 |
| LIF | 9 | 34.56 | 3.828 | 2.52 | 1.05E−26 | 2.60 | 8.75E−10 |
| SERPINB2 | 8 | 34.45 | 0.689 | 1.68 | 4.64E−07 | 4.55 | 2.23E−33 |
| SPRR1A | 8 | 29.28 | 86.666 | 1.67 | 0.00927 | 5.88 | 1.43E−14 |
| SPRR1B | 8 | 29.28 | 86.666 | 1.53 | 0.001373 | 3.48 | 1.15E−38 |
| IVL | 8 | 29.11 | 136 | 2.02 | 2.08E−11 | 2.97 | 3.49E−20 |
| DDX58 | 8 | 33.9 | 109 | 1.50 | 0.0088 | 1.55 | 2.54E−05 |
| SPRR2A | 8 | 29.28 | 86.666 | 3.57 | 4.3E−20 | 18.08 | 1.04E−152 |
| ZC3H12A | 7 | 33.56 | 4.0976 | 3.24 | 5.01E−30 | 1.92 | 3.20E−12 |
| S100A9 | 7 | 33.78 | 0.333 | 2.21 | 7.38E−35 | 2.97 | 6.39E−27 |
| PDGFB | 7 | 33.78 | 138.966 | 2.06 | 6.93E−07 | 1.95 | 3.74E−08 |
| IL7R | 7 | 33.4 | 4.91 | 1.83 | 0.013 | 2.48 | 0.0414 |
| MAP3K8 | 6 | 32.73 | 0.44 | 2.22 | 0.005 | 2.25 | 1.96E−11 |
| IKBKE | 6 | 31.98 | 3.31 | 1.71 | 0.00017 | 1.68 | 9.13E−07 |
| MAF | 3 | 30.48 | 136 | − 1.73 | 3.26E−06 | − 1.81 | 0.00755 |
| ADAM8 | 3 | 29.11 | 268 | 2.18 | 3.26E−06 | 2.33 | 3.83E−15 |
| GBP5 | 3 | 29.73 | 31.94 | 3.84 | 0.02372 | 4.26 | 4.21E−10 |
Figure 4.
Association of COVID 19 hub genes with other respiratory viral infections. A network generated by use of Cytoscape depicts the expression of COVID-19 hub genes on infection by various respiratory viruses. The green arrows indicate down-regulation, and the red arrows show up-regulation of these genes.
Discussion
Microarray meta-analysis and comparative transcriptome analysis have been useful bioinformatic approaches for maximum utilization of publicly available gene expression data16,17. Since the advent of high-throughput technologies such as microarrays and RNA-seq, researchers have performed in-depth transcriptome analyses of various biological conditions, leading to various discoveries18. Data from thousands of such experiments are being deposited in public repositories such as GEO and ArrayExpress. Selection and combinatorial analysis of such data can aid researchers in understanding the molecular mechanisms of a disease and in discovering biomarkers19–21. We observed the availability of various microarray studies related to human respiratory viral infection and sensed the opportunity to compare the effect of SARS-CoV-2 and other respiratory viral infections on the human lung transcriptome.
The comparative transcriptome analysis led to identification of genes that were altered exclusively after SARS-CoV-2 infection. Among these genes, S100 calcium-binding protein A9 (S100A9) and S100 calcium-binding protein A8 (S100A8) are calcium- and zinc-binding proteins that are elevated in inflammatory lung disorders22. S100A8 and S100A9 form a heterodimer complex called Calprotectin (CLP). Elevated levels of CLP is found in bronchoalveolar lavage fluid (BALF), serum and lung tissue of pneumonia patients23. Serum CLP could be potential biomarker for COVID-19 and further research is required to test this hypothesis24. Colony stimulating factor 2 (CSF2) is a cytokine-coding gene associated with respiratory diseases such as pulmonary alveolar proteinosis25. CSF2 (GM-CSF) is known to be pro-inflammatory cytokine produced by wide variety of cells such as macrophages, T-cells, fibroblast, tumor cells, endothelial cells, and with primary production at the inflammation site26,27. GM-CSF influences activation and proliferation of immune cells such as macrophages, monocytes, dendritic cells, neutrophils, eosinophils28. Although GM-CSF plays important role in maintaining immune homeostasis, its over-expression in lung could lead to fibrotic reactions and severe immune cell infiltrations29. Recent reports show that a) COVID-19 patients requiring intensive care unit (ICU) have increased level of GM-CSF in China and b) drugs targeting CSF2 (GM-CSF) or its receptor (such as lenzilumab, namilumab, gimsilumab, and otilimab) are being evaluated in clinical trials with COVID-19 patients13–15,30. MAS-related GPR family member X3 (MRGPRX3), a member of the mas-related/sensory neuron specific subfamily of G protein coupled receptors, is down-regulated in human airway epithelial cells exposed to smoke from electronic cigarettes31. In oral cancers, lung cancers, and head and neck cancers, C-X-C Motif chemokine ligand 14 (CXCL14) functions as a tumor suppressor; it also induces growth of prostate and breast cancers32–36. Protocadherin 7 (PCDH7) is involved in cell–cell recognition and adhesion37. Mab-21-like 4 (MAB21L4) has no known association with lung disorders or respiratory virus infections.
The PPI analysis of genes differentially expressed by SARS-CoV-2 infection identified 43 hub genes. CSF2 and S100A9 were the only hub genes to show a SARS-CoV-2-exclusive gene expression pattern. Almost all other hub genes were affected by infection of other respiratory viruses. In conclusion, both PPI analysis and comparative transcriptome analysis point to a role of CSF2 in the molecular mechanism of SARS-CoV-2 infections of human lung epithelium. The current study also highlights the exclusivity of known lung inflammation disorder genes such as S100A8 and S100A9 with respect to SARS-CoV-2 infection.
Our current study utilizes exclusively the transcriptomic data. Similar comparative analysis as well as integrative analysis with proteomics and other “omic” data will be useful as more data becomes public. The current bioinformatics approach makes use of available data to identify potential molecular targets for treatment of COVID-19. Future studies will focus on experimental validation to establish the exclusive association of CSF2, S100A8 and S100A9 with SARS-CoV-2 infections in lung epithelial cells.
Methods
RNA sequencing data analysis
The NCBI GEO database was searched for microarray or RNA-sequencing data related to SARS-CoV-2 infections of human lung epithelial cells. We evaluated two studies, GSE147507 and GSE1539706,38. GSE147507 included normal human bronchial epithelial cells subjected to mock treatment (n = 3) or to SARS-CoV-2 infection (n = 3); GSE153970 included primary human airway epithelial cultures infected with mock (n = 3) or SARS-CoV-2 (n = 3). For the purpose of validation, we also selected RNA-seq data related to SARS-CoV-2-infected human bronchial organoids [GSE150819].
Raw sequencing data related to selected samples were downloaded from Sequence Read Archive (SRA) using fastq-dump of sratoolkit v2.9.6 (http://ncbi.github.io/sra-tools/). First, raw sequencing reads were trimmed to remove adapter sequences and low-quality regions using Trim Galore! (v0.4.1) (http://www.bioinformatics.babraham.ac.uk/projects/trim_galore/). Trimmed reads were subjected to quality control analysis using FastQC (https://www.bioinformatics.babraham.ac.uk/projects/fastqc/). Tophat v2.1 was used to map trimmed raw reads to the human reference genome (hg38)39. All bam files from multiple runs related to the same samples were merged and sorted using SAMtools (Version: 1.3.1)40. Finally, raw read counts were enumerated for each gene in each sample using GTF (gene transfer file) from Ensembl [Homo.sapiens_GRCh38.82.gtf] and HTSeq-count41.
Analysis of differential expression was performed using DESeq2 according to a standard protocol (https://bioconductor.org/packages/release/bioc/vignettes/DESeq2/inst/doc/DESeq2.html)42. Genes with adj.P-value < 0.05 and absolute fold change ≥ 1.5 were considered as significantly differentially expressed. Common up-regulated and down-regulated genes from GSE147507 and GSE153970 were obtained using the Venny online tool (https://bioinfogp.cnb.csic.es/tools/venny/).
Gene ontology enrichment analyses of the common Differentially Expressed Genes (DEGs) were accomplished by use of the Database for Annotation, Visualization and Integrated Discovery (DAVID) v6.8 online tool43. Gene ontology (GO) biological processes with P-values < 0.05 and gene counts > 2 were considered as significantly enriched.
Microarray data collection and analysis
The NCBI GEO database was queried for microarray data related to SARS-CoV infections of human lung epithelial cells. A query (SARS-CoV) AND "Homo sapiens"[porgn] AND ("gse"[Filter] AND ("Expression profiling by array"[Filter])) led to 15 search results. After screening, two studies (GSE47962, GSE17400) were selected. To find microarray data related to SARS-CoV infections of human lung epithelial cells, the GEO database was queried using (((Human lung epithelium) OR (Human bronchial epithelial) AND "Homo sapiens"[porgn] AND ("gse"[Filter] AND "Expression profiling by array"[Filter]))) AND (viral infection AND ("gse"[Filter] AND "Expression profiling by array"[Filter])) AND ("gse"[Filter] AND "Expression profiling by array"[Filter]) AND ("Expression profiling by array"[Filter]). This led to 38 search results, three of which (GSE49840, GSE71766, and GSE48575) were selected for analysis. Table 3 provides sample, platform, and cell line details for all five studies.
Table 3.
Summary of publicly available, high-throughput experimental data considered in the current study.
| GEO accession number | Cell line | Virus infection studied after 24 h (along with GSM ids) | Platform considered |
|---|---|---|---|
| GSE147507 | Normal Human Bronchial Epithelial (NHBE) | SARS-CoV-2 (GSM4432381, GSM4432382, GSM4432383), Mock treatment (GSM4432378, GSM4432379, GSM4432380) | Illumina NextSeq 500 (Homo sapiens) |
| GSE153970 | Human airway epithelium (HAE) | SARS-CoV-2 (GSM4661083, GSM4661084, GSM4661085), Mock treatment (GSM4661080, GSM4661081, GSM4661082) | Illumina NovaSeq 6000 (Homo sapiens) |
| GSE47962 | Human airway epithelium (HAE) | SARS-CoV (GSM1163617, GSM1163618, GSM1163619), H1N1 (GSM1163602, GSM1163603, GSM1163604), SARS-CoV-BatSRBD (GSM1163543,GSM1163544), SARS-CoV-dORF6 (GSM1163569, GSM1163570, GSM1163571), Mock treatment (GSM1163650,GSM1163651, GSM1163652) | Agilent-014850 whole human genome microarray 4 × 44 K G4112F |
| GSE49840 | Polarized Calu3 | H7N7 (GSM1208108, GSM1208109, GSM1208110, GSM1208111), H5N1 (GSM1208139, GSM1208140, GSM1208141, GSM1208142), H3N2 (GSM1208124, GSM1208125, GSM1208126, GSM1208127), H7N9 (GSM1208076, GSM1208077, GSM1208078, GSM1208079), Mock treatment (GSM1208092, GSM1208093, GSM1208094, GSM1208095) | Agilent-039494 SurePrint G3 human GE v2 8 × 60 K microarray |
| GSE17400 | Calu3 subclone 2B4 | SARS-CoV (GSM432332, GSM432333, GSM432360), DOHV (GSM432398, GSM432399, GSM432400), Mock treatment (GSM432033, GSM432034, GSM432209) | Affymetrix human genome U133 Plus 2.0 array |
| GSE71766 | Human bronchial epithelial (BEAS-2B) | H1N1 (GSM1844862, GSM1844863, GSM1844864, GSM1844865, GSM1844866), RV16 (GSM1844907, GSM1844908, GSM1844909, GSM1844910, GSM1844911), Control (GSM1844817, GSM1844819, GSM1844820, GSM1844818, GSM1844821), H1N1 + RV16 (GSM1844952, GSM1844953, GSM1844954, GSM1844955, GSM1844956) | Affymetrix human genome U219 array |
| GSE48575 | Normal human bronchial epithelial cells (NHBECs) | H1N1pdm (GSM1181433, GSM1181434, GSM1181435), sH1N1 (GSM1181442, GSM1181443, GSM118144), Control for H1N1pdm (GSM1181427, GSM1181428, GSM1181429), Control for sH1N1(GSM1181436, GSM1181437, GSM1181438) | Illumina Human HT-12 V4.0 expression beadchip |
| GSE150819 | Human bronchial organoids | Uninfected human bronchial organoid (GSM4559193, GSM4559194, GSM4559195), SARS-CoV-2 infected human bronchial organoid (GSM4559196, GSM4559197, GSM4559198) | Illumina NovaSeq 6000 |
Since SARS-CoV-2 RNA-seq data included transcriptome profiling after 24 h of infection, in all microarray studies, we considered only samples after 24 h of viral infection.
GSE47962 included samples from human airway epithelium (HAE) cells infected with SARS-CoV, influenza virus (H1N1), or variants of SARS-CoV (SARS-dORF6 and SARS-BatSRBD)44. GSE71766 comprised human bronchial epithelial cells (BEAS-2B) infected with rhino virus (RV), influenza virus (H1N1), or both (RV + H1N1)45. Bronchial epithelial cell line 2B4 (a clonal derivative of Calu-3 cells) infected with SARS-CoV or Dhori virus (DOHV) was part of GSE1740046. GSE49840 included polarized calu-3 (cultured human airway epithelial cells) infected with human influenza virus (H3N2) or avian influenza viruses (H7N9, H5N1, and H7N7)47. GSE48575 consisted of normal human bronchial epithelial cells (NHBEC) infected with seasonal H1N1 influenza A (sH1N1) or pandemic H1N1 influenza A (H1N1pdm)48. GEO2R was used to identify differentially expressed genes for each of these studies independently12. Probes with adj. P-value < 0.05 and absolute fold change ≥ 1.5 were considered as statistically significant and were compared with DEGs of SARS-CoV-2 infection from RNA-seq data.
Protein–protein interaction analysis
STRING, a database of known or predicted protein–protein interactions (PPIs) was used to obtain interactions between genes altered on SARS-CoV-2 infection49. Output from the STRING database was uploaded to Cytoscape v3.7.2 in simple interaction format, and the Cytohubba app was employed to identify hub genes50–52. The top 50 genes were obtained separately from the PPI network based on three network parameters (closeness, degree, and betweenness) then common genes among these were selected as hub genes. We also checked the DrugBank database to determine if a drug is available to target them53.
Supplementary Information
Acknowledgements
This study was supported by UAB impact funds to UM and SV. We thank Dr. Donald Hill, of the UAB Comprehensive Cancer Center, for editing this manuscript.
Author contributions
D.S.C. and S.V. conceived, designed and supervised the research. D.S.C. performed all in silico data analysis. D.S.C. prepared figures and tables. D.S.C., S.V., U.M. and M.A. drafted and critically revised the manuscript.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors jointly supervised this work: Upender Manne and Sooryanarayana Varambally.
Contributor Information
Darshan S. Chandrashekar, Email: dshimogachandrasheka@uabmc.edu
Sooryanarayana Varambally, Email: soorya@uab.edu.
Supplementary Information
The online version contains supplementary material available at 10.1038/s41598-021-95733-w.
References
- 1.Carvalho A, Cezarotti Filho ML, Azevedo PCP, Silveira Filho RN, Barbosa FT, Rocha TJM, et al. Epidemiology, diagnosis, treatment, and future perspectives concerning SARS-COV-2: A review article. Rev Assoc Med Bras. (1992) 2020;66(3):370–374. doi: 10.1590/1806-9282.66.3.370. [DOI] [PubMed] [Google Scholar]
- 2.Nicola M, Alsafi Z, Sohrabi C, Kerwan A, Al-Jabir A, Iosifidis C, et al. The socio-economic implications of the coronavirus pandemic (COVID-19): A review. Int. J. Surg. 2020;78:185–193. doi: 10.1016/j.ijsu.2020.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Singh SK. Respiratory viral infections. Semin. Respir. Crit. Care Med. 2016;37(4):485–486. doi: 10.1055/s-0036-1584802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Afrough B, Dowall S, Hewson R. Emerging viruses and current strategies for vaccine intervention. Clin. Exp. Immunol. 2019;196(2):157–166. doi: 10.1111/cei.13295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wang C, Li W, Drabek D, Okba NMA, van Haperen R, Osterhaus A, et al. A human monoclonal antibody blocking SARS-CoV-2 infection. Nat. Commun. 2020;11(1):2251. doi: 10.1038/s41467-020-16256-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Blanco-Melo D, Nilsson-Payant BE, Liu WC, Uhl S, Hoagland D, Moller R, et al. Imbalanced host response to SARS-CoV-2 drives development of COVID-19. Cell. 2020;181(5):1036–1045e9. doi: 10.1016/j.cell.2020.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Messina F, Giombini E, Agrati C, Vairo F, Ascoli Bartoli T, Al Moghazi S, et al. COVID-19: Viral-host interactome analyzed by network based-approach model to study pathogenesis of SARS-CoV-2 infection. J. Transl. Med. 2020;18(1):233. doi: 10.1186/s12967-020-02405-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kumar N, Mishra B, Mehmood A, Mohammad A, Mukhtar MS. Integrative network biology framework elucidates molecular mechanisms of SARS-CoV-2 pathogenesis. iScience. 2020;23(9):101526. doi: 10.1016/j.isci.2020.101526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yue Z, Zhang E, Xu C, Khurana S, Batra N, Dang SDH, et al. PAGER-CoV: A comprehensive collection of pathways, annotated gene-lists and gene signatures for coronavirus disease studies. Nucleic Acids Res. 2020;49:D589–D599. doi: 10.1093/nar/gkaa1094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhang N, Wang L, Deng X, Liang R, Su M, He C, et al. Recent advances in the detection of respiratory virus infection in humans. J. Med. Virol. 2020;92(4):408–417. doi: 10.1002/jmv.25674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Parkinson H, Kapushesky M, Shojatalab M, Abeygunawardena N, Coulson R, Farne A, et al. ArrayExpress—A public database of microarray experiments and gene expression profiles. Nucleic Acids Res. 2007;35(database issue):D747–D750. doi: 10.1093/nar/gkl995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Barrett T, Wilhite SE, Ledoux P, Evangelista C, Kim IF, Tomashevsky M, et al. NCBI GEO: Archive for functional genomics data sets–update. Nucleic Acids Res. 2013;41(database issue):D991–D1005. doi: 10.1093/nar/gks1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mehta P, Porter JC, Manson JJ, Isaacs JD, Openshaw PJM, McInnes IB, et al. Therapeutic blockade of granulocyte macrophage colony-stimulating factor in COVID-19-associated hyperinflammation: Challenges and opportunities. Lancet Respir. Med. 2020;8:822–830. doi: 10.1016/S2213-2600(20)30267-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lang FM, Lee KM, Teijaro JR, Becher B, Hamilton JA. GM-CSF-based treatments in COVID-19: Reconciling opposing therapeutic approaches. Nat. Rev. Immunol. 2020;23:1–8. doi: 10.1038/s41577-020-0357-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Temesgen Z, Assi M, Shweta FNU, Vergidis P, Rizza SA, Bauer PR, et al. GM-CSF neutralization with lenzilumab in severe COVID-19 pneumonia: A case-cohort study. Mayo Clin. Proc. 2020;95(11):2382–2394. doi: 10.1016/j.mayocp.2020.08.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ramasamy A, Mondry A, Holmes CC, Altman DG. Key issues in conducting a meta-analysis of gene expression microarray datasets. PLoS Med. 2008;5(9):e184. doi: 10.1371/journal.pmed.0050184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hamid, J.S., Hu, P., Roslin, N.M., Ling, V., Greenwood, C.M., & Beyene, J. Data integration in genetics and genomics: Methods and challenges. Hum. Genomics Proteomics. 2009 (2009). [DOI] [PMC free article] [PubMed]
- 18.Cahan P, Rovegno F, Mooney D, Newman JC, St Laurent G, McCaffrey TA. Meta-analysis of microarray results: Challenges, opportunities, and recommendations for standardization. Gene. 2007;401(1–2):12–18. doi: 10.1016/j.gene.2007.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chen JA, Yu Y, Xue C, Chen XL, Cui GY, Li J, et al. Low microRNA-139 expression associates with poor prognosis in patients with tumors: A meta-analysis. Hepatobiliary Pancreat. Dis. Int. 2019;18(4):321–331. doi: 10.1016/j.hbpd.2018.09.016. [DOI] [PubMed] [Google Scholar]
- 20.Sherafatian M, Abdollahpour HR, Ghaffarpasand F, Yaghmaei S, Azadegan M, Heidari M. MicroRNA expression profiles, target genes, and pathways in intervertebral disk degeneration: A meta-analysis of 3 microarray studies. World Neurosurg. 2019;126:389–397. doi: 10.1016/j.wneu.2019.03.120. [DOI] [PubMed] [Google Scholar]
- 21.Huang W, Ran R, Shao B, Li H. Prognostic and clinicopathological value of PD-L1 expression in primary breast cancer: A meta-analysis. Breast Cancer Res. Treat. 2019;178(1):17–33. doi: 10.1007/s10549-019-05371-0. [DOI] [PubMed] [Google Scholar]
- 22.Gomes LH, Raftery MJ, Yan WX, Goyette JD, Thomas PS, Geczy CL. S100A8 and S100A9-oxidant scavengers in inflammation. Free Radic. Biol. Med. 2013;58:170–186. doi: 10.1016/j.freeradbiomed.2012.12.012. [DOI] [PubMed] [Google Scholar]
- 23.Kotsiou OS, Papagiannis D, Papadopoulou R, Gourgoulianis KI. Calprotectin in lung diseases. Int. J. Mol. Sci. 2021;22(4):1706. doi: 10.3390/ijms22041706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mahler M, Meroni PL, Infantino M, Buhler KA, Fritzler MJ. Circulating calprotectin as a biomarker of COVID-19 severity. Expert Rev. Clin. Immunol. 2021;17(5):431–443. doi: 10.1080/1744666X.2021.1905526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ito M, Nakagome K, Ohta H, Akasaka K, Uchida Y, Hashimoto A, et al. Elderly-onset hereditary pulmonary alveolar proteinosis and its cytokine profile. BMC Pulm. Med. 2017;17(1):40. doi: 10.1186/s12890-017-0382-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Griffin JD, Cannistra SA, Sullivan R, Demetri GD, Ernst TJ, Kanakura Y. The biology of GM-CSF: Regulation of production and interaction with its receptor. Int. J. Cell Cloning. 1990;8(Suppl 1):35–44. doi: 10.1002/stem.5530080705. [DOI] [PubMed] [Google Scholar]
- 27.Shiomi A, Usui T. Pivotal roles of GM-CSF in autoimmunity and inflammation. Mediators Inflamm. 2015;2015:568543. doi: 10.1155/2015/568543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hamilton JA. Colony-stimulating factors in inflammation and autoimmunity. Nat. Rev. Immunol. 2008;8(7):533–544. doi: 10.1038/nri2356. [DOI] [PubMed] [Google Scholar]
- 29.Xing Z, Ohkawara Y, Jordana M, Graham F, Gauldie J. Transfer of granulocyte-macrophage colony-stimulating factor gene to rat lung induces eosinophilia, monocytosis, and fibrotic reactions. J. Clin. Invest. 1996;97(4):1102–1110. doi: 10.1172/JCI118503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Huang C, Wang Y, Li X, Ren L, Zhao J, Hu Y, et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395(10223):497–506. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Solleti SK, Bhattacharya S, Ahmad A, Wang Q, Mereness J, Rangasamy T, et al. MicroRNA expression profiling defines the impact of electronic cigarettes on human airway epithelial cells. Sci. Rep. 2017;7(1):1081. doi: 10.1038/s41598-017-01167-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ozawa S, Kato Y, Komori R, Maehata Y, Kubota E, Hata R. BRAK/CXCL14 expression suppresses tumor growth in vivo in human oral carcinoma cells. Biochem. Biophys. Res. Commun. 2006;348(2):406–412. doi: 10.1016/j.bbrc.2006.07.070. [DOI] [PubMed] [Google Scholar]
- 33.Augsten M, Hagglof C, Olsson E, Stolz C, Tsagozis P, Levchenko T, et al. CXCL14 is an autocrine growth factor for fibroblasts and acts as a multi-modal stimulator of prostate tumor growth. Proc. Natl. Acad. Sci. U S A. 2009;106(9):3414–3419. doi: 10.1073/pnas.0813144106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ozawa S, Kato Y, Ito S, Komori R, Shiiki N, Tsukinoki K, et al. Restoration of BRAK/CXCL14 gene expression by gefitinib is associated with antitumor efficacy of the drug in head and neck squamous cell carcinoma. Cancer Sci. 2009;100(11):2202–2209. doi: 10.1111/j.1349-7006.2009.01281.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tessema M, Klinge DM, Yingling CM, Do K, Van Neste L, Belinsky SA. Re-expression of CXCL14, a common target for epigenetic silencing in lung cancer, induces tumor necrosis. Oncogene. 2010;29(37):5159–5170. doi: 10.1038/onc.2010.255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Augsten M, Sjoberg E, Frings O, Vorrink SU, Frijhoff J, Olsson E, et al. Cancer-associated fibroblasts expressing CXCL14 rely upon NOS1-derived nitric oxide signaling for their tumor-supporting properties. Cancer Res. 2014;74(11):2999–3010. doi: 10.1158/0008-5472.CAN-13-2740. [DOI] [PubMed] [Google Scholar]
- 37.Nakamura H, Nakashima T, Hayashi M, Izawa N, Yasui T, Aburatani H, et al. Global epigenomic analysis indicates protocadherin-7 activates osteoclastogenesis by promoting cell-cell fusion. Biochem. Biophys. Res. Commun. 2014;455(3–4):305–311. doi: 10.1016/j.bbrc.2014.11.009. [DOI] [PubMed] [Google Scholar]
- 38.Vanderheiden A, Ralfs P, Chirkova T, Upadhyay AA, Zimmerman MG, Bedoya S, et al. Type I and type III interferons restrict SARS-CoV-2 infection of human airway epithelial cultures. J. Virol. 2020;94(19):e00985. doi: 10.1128/JVI.00985-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kim D, Pertea G, Trapnell C, Pimentel H, Kelley R, Salzberg SL. TopHat2: Accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 2013;14(4):R36. doi: 10.1186/gb-2013-14-4-r36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Li H. A statistical framework for SNP calling, mutation discovery, association mapping and population genetical parameter estimation from sequencing data. Bioinformatics. 2011;27(21):2987–2993. doi: 10.1093/bioinformatics/btr509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Anders S, Pyl PT, Huber W. HTSeq—A Python framework to work with high-throughput sequencing data. Bioinformatics. 2015;31(2):166–169. doi: 10.1093/bioinformatics/btu638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15(12):550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Dennis G, Jr, Sherman BT, Hosack DA, Yang J, Gao W, Lane HC, et al. DAVID: Database for annotation, visualization, and integrated discovery. Genome Biol. 2003;4(5):P3. doi: 10.1186/gb-2003-4-5-p3. [DOI] [PubMed] [Google Scholar]
- 44.Mitchell HD, Eisfeld AJ, Sims AC, McDermott JE, Matzke MM, Webb-Robertson BJ, et al. A network integration approach to predict conserved regulators related to pathogenicity of influenza and SARS-CoV respiratory viruses. PLoS ONE. 2013;8(7):e69374. doi: 10.1371/journal.pone.0069374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kim TK, Bheda-Malge A, Lin Y, Sreekrishna K, Adams R, Robinson MK, et al. A systems approach to understanding human rhinovirus and influenza virus infection. Virology. 2015;486:146–157. doi: 10.1016/j.virol.2015.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yoshikawa T, Hill TE, Yoshikawa N, Popov VL, Galindo CL, Garner HR, et al. Dynamic innate immune responses of human bronchial epithelial cells to severe acute respiratory syndrome-associated coronavirus infection. PLoS ONE. 2010;5(1):e8729. doi: 10.1371/journal.pone.0008729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Josset L, Zeng H, Kelly SM, Tumpey TM, Katze MG. Transcriptomic characterization of the novel avian-origin influenza A (H7N9) virus: specific host response and responses intermediate between avian (H5N1 and H7N7) and human (H3N2) viruses and implications for treatment options. MBio. 2014;5(1):e01102–e1113. doi: 10.1128/mBio.01102-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Paquette SG, Banner D, le Chi TB, Leomicronn AJ, Xu L, Ran L, et al. Pandemic H1N1 influenza A directly induces a robust and acute inflammatory gene signature in primary human bronchial epithelial cells downstream of membrane fusion. Virology. 2014;448:91–103. doi: 10.1016/j.virol.2013.09.022. [DOI] [PubMed] [Google Scholar]
- 49.von Mering C, Jensen LJ, Snel B, Hooper SD, Krupp M, Foglierini M, et al. STRING: Known and predicted protein-protein associations, integrated and transferred across organisms. Nucleic Acids Res. 2005;33(Database issue):D433–D437. doi: 10.1093/nar/gki005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, et al. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13(11):2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Su G, Morris JH, Demchak B, Bader GD. Biological network exploration with Cytoscape 3. Curr. Protoc. Bioinform. 2014;47:8131–8224. doi: 10.1002/0471250953.bi0813s47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chin CH, Chen SH, Wu HH, Ho CW, Ko MT, Lin CY. cytoHubba: Identifying hub objects and sub-networks from complex interactome. BMC Syst. Biol. 2014;8(Suppl 4):S11. doi: 10.1186/1752-0509-8-S4-S11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wishart DS, Knox C, Guo AC, Cheng D, Shrivastava S, Tzur D, et al. DrugBank: A knowledgebase for drugs, drug actions and drug targets. Nucleic Acids Res. 2008;36(Database issue):D901–D906. doi: 10.1093/nar/gkm958. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.