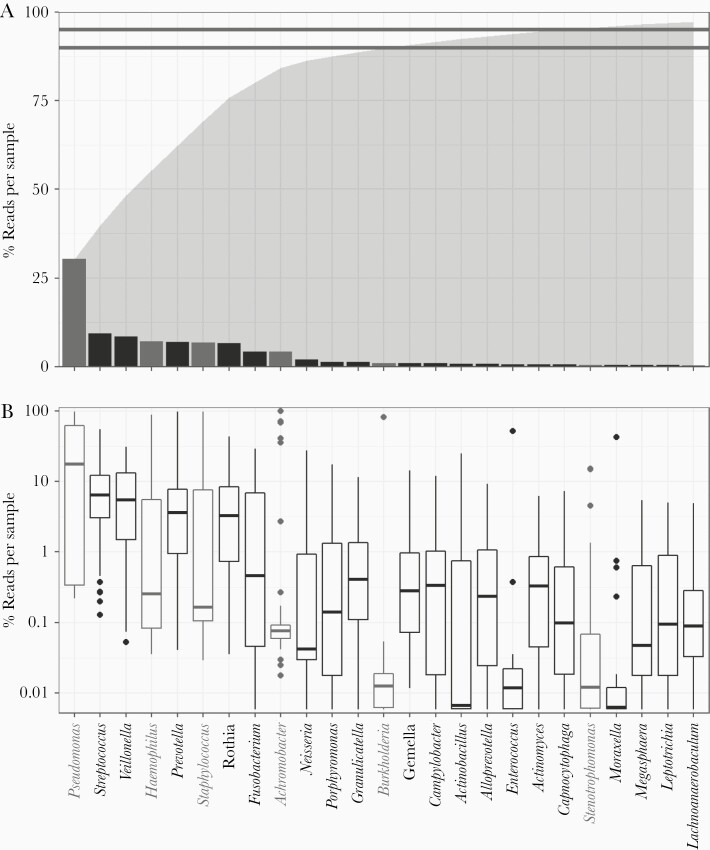
Figure 1.
Cystic fibrosis (CF) lung microbiomes are dominated by oral anaerobes and opportunistic pathogens We analyzed CF sputum expectorate (n = 77) using 16S sequencing and an in-house QIIME 2-based bioinformatics pipeline to resolve strain-level Operational Taxonomic Units (OTUs). Samples were rarefied to 17 000 reads. We identified 217 OTUs across 59 genera and at least 81 species. Overall, we found that CF sputum samples were dominated by oral anaerobes and opportunistic pathogens. A, Sequences mapped to 14 genera comprised 90% (lower line) of the total reads obtained; 95% (upper line) of all reads mapped to 21 genera. Total cumulative read fraction is represented in shaded region. Recognized CF pathogens are as follows: Pseudomonas, Haemophilus, Staphylococcus, Achromobacter, Burkholderia, and Stenotrophomonas. Pseudomonas was the most prevalent genus, followed by Streptococcus and Veillonella. B, Binning reads by sample shows variation in relative abundance. Pseudomonas comprised >10% of reads in the majority of our samples. While over 6% of the total reads mapped to Achromobacter, only 4 samples comprised >10% Achromobacter.