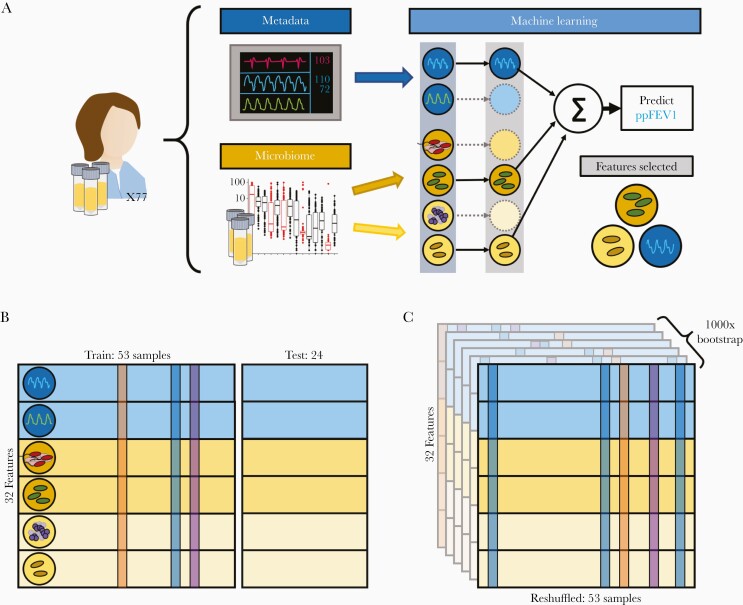
Figure 4.
Machine learning overview. Machine learning models were trained on different input data tables using varying data resampling methods. A, Features were categorized by information source (microbiome or patient metadata). The 16S data was further split into pathogens and other taxa in agreement with Figure 2. We used ElasticNet regularization to select informative features that predict ppFEV1. B, We randomly selected 24 patient samples to withhold as a test set and trained our models on the remaining 53 samples. To assess overfitting, we used leave-one-out cross-validation on our training set. C, We additionally implemented 1000-fold bootstrap resampling to assess the robustness of our model fits. Abbreviation: ppFEV1, percent predicted forced expiratory volume in 1 second.