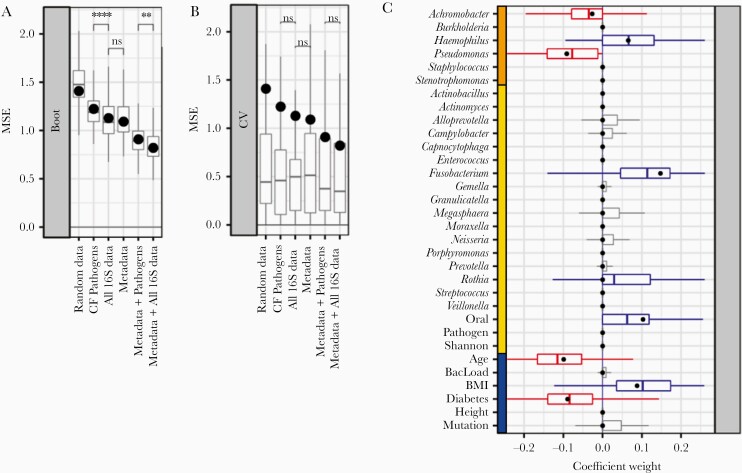
Figure 5.
Bootstrapped ElasticNet-identified predictors of lung function. Machine learning models were trained using varying input datasets. A, 1000-fold bootstrapping and (B) leave-one-out cross-validation (LOOCV) were used to generate prediction error (MSE) ranges across feature subsets. Models trained on all of the data showed lower error compared to other feature subsets. Adding 16S pathogen quantitation decreased model error. Models trained on all 16S data outperformed models using only 16S pathogen quantitation (P < .01, t test). Regardless of input features, models trained on the full sample set (black points) were greater than median LOOCV MSEs (boxplots). C, Coefficient ranges for train/test (black points) and bootstrapped models (boxplots) trained on standardized input datasets (blue, metadata; orange, 16S pathogens; yellow, 16S other taxa) show consistency between both machine learning strategies. Both cases selected Pseudomonas and Achromobacter as negative predictors. Abbreviations: BMI, body mass index; CF, cystic fibrosis; MSE, mean squared error; ns, not significant. **P < .01; ****P < .0001.