Figure 6. Iterative refinement of cell identity using multiple single-cell modalities from the mouse primary motor cortex.
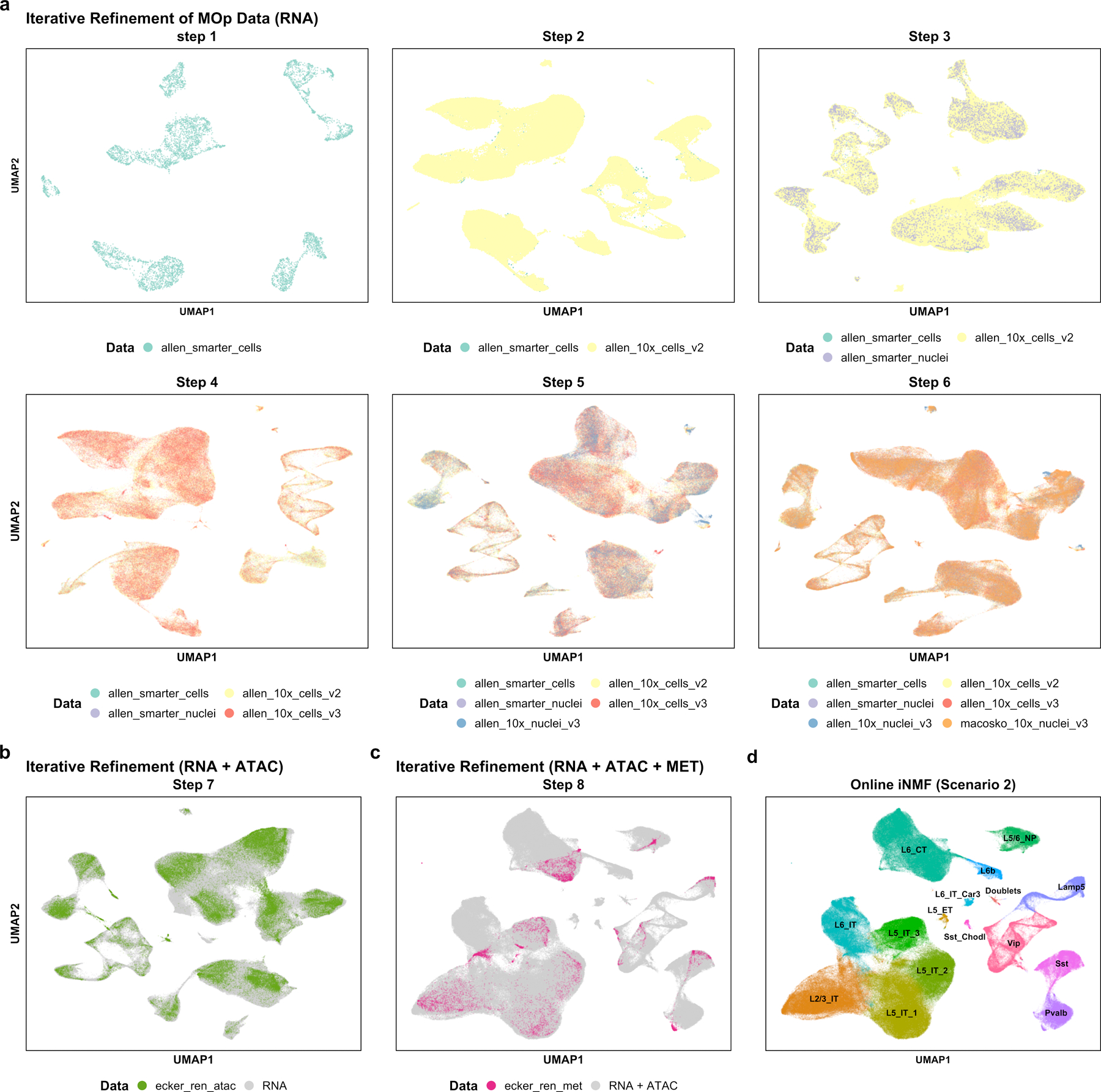
We integrated four scRNA-seq datasets, two snRNA-seq datasets, one snATAC-seq dataset and one snmC-seq dataset . a, Sequential integration of six scRNA-seq datasets (scenario 2). Each panel shows a UMAP plot using cell factors obtained after adding an additional dataset. b, UMAP plot of cell factors obtained by adding snATAC-seq to the latent space learned from six RNA datasets in a (scenario 2). c, UMAP plot of cell factors obtained by adding DNA methylation data (snmC-seq, abbreviated “MET”) to the latent space learned from the seven datasets shown in b (scenario 2). d, Clusters obtained using the cell factor loadings of all eight aligned datasets. The clusters were named using marker genes from Tasic et al28.
