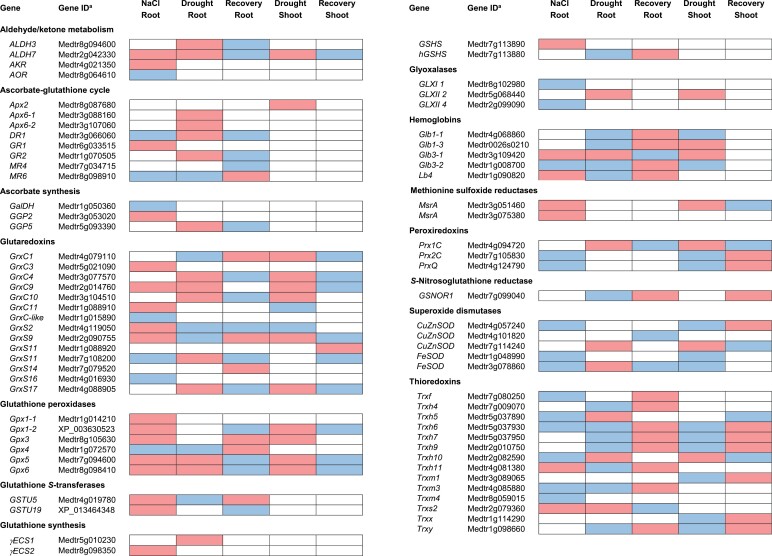
Fig. 4.
Expression profile of genes involved in redox homeostasis in the roots and shoots of Medicago truncatula plants exposed to drought or salt stress. Gene up-regulation (>two-fold) and down-regulation (<0.5-fold) are indicated in red and blue, respectively. Gene IDs are given according to the M. truncatula genome v4.0. Transcriptomic data of roots and shoots under drought stress were retrieved from the Gene Expression Atlas (https://mtgea.noble.org/v3/) and those of salt stress in the roots were published by Li et al., (2009). Gene sequences were obtained from Legume IP (http://plantgrn.noble.org/LegumeIP/gdp/), except for Gpx1-2 and GSTU19, which were obtained from GenBank. The probes used to determine gene expression profiles are listed in Table 1. Abbreviations: AKR, aldo-keto reductase; ALDH, aldehyde dehydrogenase; Apx, ascorbate peroxidase; AOR, alkenal/one oxidoreductase; DR, dehydroascorbate reductase; γECS, γ-glutamylcysteine synthetase; GalDH, L-galactono-1,4-lactone dehydrogenase; GGP, GDP-L-galactose phosphorylase; Glb, phytoglobin; GLX, glyoxalase; Gpx, glutathione peroxidase; GR, glutathione reductase; Grx, glutaredoxin; GSHS, glutathione synthetase; GSNOR, S-nitrosoglutathione reductase; GSTU, glutathione S-transferase tau family; hGSHS, homoglutathione synthetase; Lb, leghemoglobin; MR, monodehydroascorbate reductase; Msr, methionine sulfoxide reductase; Prx, peroxiredoxin; SOD, superoxide dismutase; Trx, thioredoxin.